Journal of Human Evolution
Volume 172, November 2022, 103258
Sedimentary ancient DNA metabarcoding as a tool for assessing prehistoric plant use at the Upper Paleolithic cave site Aghitu-3, Armenia
Author links open overlay panelAnneke T.M.ter SchureaPersonEnvelopeAngela A.BruchbAndrew W.KandelcBorisGasparyandRainer W.BussmanneAnne K.BrystingaHugo J.de BoerfSanneBoessenkoolaPersonEnvelope
Highlights
- • We successfully recovered plant DNA from Upper Paleolithic layers at Aghitu-3 Cave.
- • Detailed assessment of pollen and plant DNA shows a diverse array of plant uses.
- • Plant DNA reflects periods of human occupation and provides evidence for plant use.
- • Detected taxa support earlier reports of prehistoric plant use as medicine and dye.
Abstract
The places in which people live, sleep, prepare food, and undertake other activities—known variably as homes, residential sites, living sites, and domestic spaces—play a key role in the emergence and evolution of modern human culture. The dynamic influence of domestic spaces began early in human evolutionary history, during the Paleolithic/Stone Age. Drawing on examples from Africa and western Eurasia, this article explores aspects of the changing social and cultural significance of domestic spaces throughout this time using several lines of evidence: repeated site visitation, behavioral structuring of living spaces, and information gained by dissecting palimpsest records. With the development of pyrotechnology, living sites become hearth-centered domestic spaces that provided a common hub for activities. Through time the activities around hearths increased in their complexity and diversity. The parsing of palimpsest records by archaeologists also reveals changes in the nature, variety, and intensity of on-site activities through time, indicating shifts in site function and the spatial expression of cultural norms. Archaeological evidence shows that the entwined development of domestic spaces and human cultural activities was gradual, albeit nonlinear from the Lower Paleolithic through the Upper Paleolithic/later Middle Stone Age. In this process, domestic spaces emerged as common arenas of opportunity for social interaction and knowledge transmission, qualities that may have contributed to and enhanced the development of cumulative culture in Paleolithic society.
Introduction
Many animals modify their environment to make safe places for sleeping or rearing their young, such as digging a burrow or building a nest. Examples among mammals include farrowing by wild female pigs and nest building by voles, packrats, and bears (Vaughan, 1990; Mayer et al., 2002; Miller et al., 2017; Zhong et al., 2022). Corresponding structures built by humans have a much broader range of uses that may include child rearing, food preparation and sharing, storage, tool manufacture, and maintaining social bonds through conversation and physical contact. Human habitations—specifically domestic spaces—also function as a theater for the performance, reenactment, transmission, and acquisition of cultural knowledge and skills (Kent, 1984; Gamble, 1999; Clark and Ranlett, 2022).
As nodes of information sharing, domestic spaces increasingly became places where culture could be perpetuated (Kuhn et al., 2018; Kuhn and Stiner, 2019). Without these spaces, the propagative engine of cumulative culture would have remained very weak in human evolution. A niche-constructing dynamic (Laland et al., 2000; Odling-Smee et al., 2003) exerted reciprocal influences between human ancestors and domestic spaces (Dunbar, 2003; Maher and Conkey, 2019). Whereas many readers may agree with all or parts of this claim in principle, making the case with scientific evidence requires that we deal effectively with the fuzziness of Paleolithic records (Stiner, 2021).
The social interactions and knowledge transmission that underlie cumulative culture are contingent on the proximity among individuals. Domestic places thus emerged as common arenas of opportunity. Here we explore evidence about the evolution of human culture in the context of domestic spaces. We begin with a closer look at the core functional properties of domestic spaces as they pertain to earlier human lifeways, and then examine the archaeological evidence for trends. These trends include the repeated visitation of certain locations, the use of fire, and the decoration, structural elaboration, and organization of living spaces. This evidence seldom comes in neat, clearly interpretable packages. Rather, we must find ways to make greater sense of palimpsest phenomena as well as the rare ‘smoking guns’ in the archaeological record. Archaeologists can learn a lot from artifact assemblages even if they have become somewhat jumbled or temporally compressed. Because domestic spaces are subject to destructive postdepositional processes and not easily interpreted, we require robust methodologies to extract meaningful information about what transpired in these locations (Clark and Gingerich, 2022).
In this review, we summarize evidence for the transformation of these spaces from >3 ma to ∼30 ka in western Eurasia and Africa. Patterns detected within these time-averaged records are cumulative and thus reflect habitual actions that traverse time, improving our understanding of the function of domestic spaces and evolution of human behaviors in relationship to those spaces in the past. We begin by exploring the concept of domestic space and its role in the development of cumulative culture. Next, we review some important changes seen in the archaeological record across five roughly defined intervals. Finally, we highlight methods to extract additional information by dissecting palimpsest records, information that cannot be had from single-component records.
Section snippets
Domestic space and its role in cumulative culture
The phenomenon of domestic space has a deep history in human evolution. Early hominins established domestic spaces to satisfy their daily physical needs, albeit in relatively straightforward ways. The defining feature of recent and ancient (Pleistocene) human domestic spaces is food preparation and consumption, usually assisted by technology. Based on prior literature and early archaeological patterns, Stiner (2021) defines domestic spaces as “widely repeated interaction nodes that involve food
Notable shifts in the character of domestic spaces
The major changes in the use of domestic space described below are broadly related to the amount of time and effort hominins were expending to create and maintain a habitable space. In the African and western Eurasian records, this notably includes the repeated visitation of sites, use of fire, occasional management of waste, structural modifications, as well as the development of symbolic elaborations with, and within, a space. Here we describe what is known archaeologically about the
Unpacking palimpsests for more clues
In the preceding narrative, we showed how Paleolithic living spaces increasingly supported a wider range of purposes and tasks with time. Much of this evidence comes from the overt spatial structuring of features, artifacts, and waste materials. In this section, we try to unpack some fuzzier structural aspects of early Paleolithic records to further understand the socioeconomic dynamics that shaped them. This is challenging due to time-dependent taphonomic processes and the fact that many
Conclusions
We have endeavored in this article to examine human behavioral shifts and transitions over large spatial and temporal scales as manifested in domestic spaces. Some of these changes emerged well before the dawn of so-called modern human behavior (Wadley, 2001; Nowell, 2010; Ames et al., 2013). With time, the places where hominins met their basic survival needs increasingly became central hubs for a greater diversity of tasks. With these changes came the possibility for more complex and
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
We wish to thank the three anonymous reviewers whose comments helped us to significantly improve this manuscript.
References (304)
- P. Goldberg et al.
New evidence on Neandertal use of fire: Examples from Roc de Marsal and Pech de l'Azé IV
Quat. Int.
(2012)
- S. Gaudzinski-Windheuser
The public and private use of space in Magdalenian societies: Evidence from Oelknitz 3, LOP (Thuringia, Germany)
J. Anthropol. Archaeol.
(2015)
- M. García-Diez et al.
Uranium series dating reveals a long sequence of rock art at Altamira Cave (Santillana del Mar, Cantabria)
J. Archaeol. Sci.
(2013)
- N. Galanidou
Patterns in caves: Foragers, horticulturists, and the use of space
J. Anthropol. Archaeol.
(2000)
- O. Fuentes et al.
An approach to Palaeolithic networks: The question of symbolic territories and their interpretation through Magdalenian art
Quat. Int.
(2019)
- V. Feruglio et al.
Rock art, performance and Palaeolithic cognitive systems: The example of the Grand Panel palimpsest of Cussac Cave, Dordogne, France
J. Anthropol. Archaeol.
(2019)
- M. Domínguez-Rodrigo et al.
The spatial patterning of the social organization of modern foraging Homo sapiens: A methodological approach for understanding social organization in prehistoric foragers
Palaeogeogr. Palaeoclimatol. Palaeoecol.
(2017)
- M. Domínguez-Rodrigo et al.
A spatial analysis of stone tools and fossil bones at FLK Zinj 22 and PTK I (Bed I, Olduvai Gorge, Tanzania) and its bearing on the social organization of early humans
Palaeogeogr. Palaeoclimatol. Palaeoecol.
(2017)
- L. Demay et al.
Mammoths used as food and building resources by Neanderthals: Zooarchaeological study applied to layer 4, Molodova I (Ukraine)
Quat. Int.
(2012)
- F. d'Errico et al.
The origin and evolution of sewing technologies in Eurasia and North America
J. Hum. Evol.
(2018)
- F. d'Errico et al.
Earliest evidence of personal ornaments associated with burial: The Conus shells from Border Cave
J. Hum. Evol.
(2016)
- P.C. Dawson
Space syntax analysis of Central Inuit snow houses
J. Anthropol. Archaeol.
(2002)
- A.E. Clark
Using spatial context to identify lithic selection behaviors
J. Archaeol. Sci. Rep.
(2019)
- C.H. Chiang
Houses” in the Wansan Society, Neolithic Taiwan
J. Anthropol. Archaeol.
(2015)
- R.N. Carmody et al.
The energetic significance of cooking
J. Hum. Evol.
(2009)
- D. Cabanes et al.
Phytolith evidence for hearths and beds in the late Mousterian occupations of Esquilleu Cave (Cantabria, Spain)
J. Archaeol. Sci.
(2010)
- M. Brenet et al.
The Late Solutrean open-air site of Landry (Aquitaine, France). A preliminary spatio-temporal analysis
Quat. Int.
(2018)
- R. Bourrillon et al.
A new Aurignacian engraving from Abri Blanchard, France: Implications for understanding Aurignacian graphic expression in Western and Central Europe
Quat. Int.
(2018)
- C. Bourdier
Rock art and social geography in the Upper Paleolithic: Contribution to the socio-cultural function of the Roc-aux-Sorciers rock-shelter (Angles-sur-l'Anglin, France) from the viewpoint of its sculpted frieze
J. Anthropol. Archaeol.
(2013)
- R. Blasco et al.
What happens around a fire: Faunal processing sequences and spatial distribution at Qesem Cave (300 ka), Israel
Quat. Int.
(2016)
- D.W. Bird et al.
Variability in the organization and size of hunter-gatherer groups: Foragers do not live in small-scale societies
J. Hum. Evol.
(2019)
- J. Birch et al.
Navigating ancestral landscapes in the Northern Iroquoian world
J. Anthropol. Archaeol.
(2015)
- İ. Baykara et al.
The Middle Paleolithic occupations of Üçağızlı II Cave (Hatay, Turkey): Geoarcheological and archeological perspectives
J. Archaeol. Sci. Rep.
(2015)
- O. Bar-Yosef et al.
Who were the makers of the Chatelperronian culture?
J. Hum. Evol.
(2010)
- O. Bar-Yosef et al.
The dating of the Upper Paleolithic layers in Kebara Cave, Mt Carmel
J. Archaeol. Sci.
(1996)
- G. Bailey
Time perspectives, palimpsests and the archaeology of time
J. Anthropol. Archaeol.
(2007)
- W. Archer et al.
What is 'in situ'? A reply to Harmand et al. (2015)
J. Hum. Evol.
(2020)
- V. Aldeias et al.
Evidence for Neandertal use of fire at Roc de Marsal (France)
J. Archaeol. Sci.
(2012)
- D.S. Adler et al.
Patterns of spatial organization and land use during the Eemian interglacial in the Rhineland: New data from Wallertheim, Germany
Eurasian Prehistory
(2003)
- T. Akazawa et al.
Neanderthal infant burial from the Dederiyeh Cave in Syria
Paléorient
(1995)
- T. Akazawa et al.
New discovery of a Neanderthal child burial from the Dederiyeh Cave in Syria
Paléorient
(1999)
- N. Alperson-Afil et al.
Differential use of space in the Neanderthal site of Amud Cave, Israel
Eurasian Prehistory
(2005)
- N. Alperson-Afil et al.
Spatial organization of hominin activities at Gesher Benot Ya'aqov, Israel
Science
(2009)
- C.J.H. Ames et al.
Why we need an alternative approach to the study of modern human behavior
Can. J. Archaeol.
(2013)
- L. Anderson et al.
Relier des espaces, construire de nouveaux réseaux: aux origines du Protoaurignacien et des débuts du Paléolithique supérieur en Europe occidentale
- M. Aubert et al.
Pleistocene cave art from Sulawesi, Indonesia
Nature
(2014)
- L.R. Backwell et al.
New excavations at Border Cave, KwaZulu-Natal, South Africa
J. Field Archaeol.
(2018)
- G. Bailey et al.
Caves, palimpsests and dwelling spaces: Examples from the Upper Palaeolithic of south-east Europe
World Archaeol.
(2009)
- R. de Balbín-Behrmann et al.
The Palaeolithic art of Tito Bustillo cave (Asturias, Spain) in its archaeological context
Quat. Int.
(2017)
- L.S. Barham
Recent research on the Middle Stone Age at Mumbwa Caves, Central Zambia
- O. Bar-Yosef et al.
The excavations in Kebara Cave, Mt. Carmel
Curr. Anthropol.
(1992)
- R. Barkai et al.
Uranium series dates from Qesem Cave, Israel, and the end of the Lower Paleolithic
Nature
(2003)
- R.G. Bednarik
Pleistocene palaeoart of Africa
Arts
(2013)
- R. Bégouën et al.
Parietal art and archaeological context: Activities of the Magdalenians in the cave at Tuc d’Audoubert
- F. Berna et al.
Microstratigraphic evidence of in situ fire in the Acheulean strata of Wonderwerk Cave, Northern Cape province, South Africa
Proc. Natl. Acad. Sci. USA
(2012)
- L.R. Binford
- L.R. Binford
Researching ambiguity: Frames of reference and site structure
- D. Borić
‘Deep time’ metaphor: Mnemonic and apotropaic practices at Lepenski Vir
J. Soc. Archaeol.
(2003)
- D. Borić
The house between grand narrative and microhistory: A house society in the Balkans
There are more references available in the full text version of this article.
Keywords
SedaDNA
Metabarcoding
Palynological analysis
Cave sediments
Armenian Highlands
1. Introduction
Analyses of preserved botanical remains yield excitingly detailed insights into the integral role that plants played in prehistoric human life, as plants provide food, medicine, raw materials, and fuel (Shipley and Kindscher, 2016; Hardy, 2018, 2019). Evidence for the Paleolithic human diet gleaned from plant fragments, phytoliths, and microfossils, as well as biomarkers from food preparation tools and dental calculus, has shifted the assumption of a largely animal-based diet to one that included a range of plants (Revedin et al., 2010; Power et al., 2018; Wadley et al., 2020). Specifically, DNA, chemical biomarkers, and starch grains extracted from dental calculus at several Neanderthal sites indicate ingestion of plant foods (Hardy et al., 2012; Salazar-García et al., 2013; Weyrich et al., 2017; Power et al., 2018). Furthermore, starch grains recovered from ∼30,000-year-old grinding tools at several locations in Europe suggest widespread vegetal food processing by hunter-gatherers, and possibly the production of flour (Revedin et al., 2010). Plants also provided fuel for fires and key raw materials (Albert et al., 2000; Allué et al., 2012; Goldberg et al., 2012), and were possibly used for medicine (Hardy, 2019), as well as early production of textiles (Kvavadze et al., 2009). Nevertheless, survival of identifiable plant remains is rare at Paleolithic sites, and our current knowledge about past human plant use is limited by the degradation of plant remains, by the ability to recover those remains, and by the availability of methods and technologies for plant detection and identification.
Developments in sedimentary ancient DNA (sedaDNA) methods and protocols have allowed the recovery of DNA from a variety of sedimentary contexts, including cave sediments (Hofreiter et al., 2003; Willerslev, 2003; Haile et al., 2007). Through such analyses we can identify organisms in the absence of any visible remains in cave environments, as illustrated by studies that successfully retrieved sedaDNA in caves originating from animals, including birds and mammals as well as hominins (Willerslev, 2003; Hofreiter et al., 2003; Haile et al., 2007; Slon et al., 2017; Zhang et al., 2020; Vernot et al., 2021). Notably, recent sedaDNA analyses of cave sites in Europe, Siberia, and the Tibetan Plateau have reconstructed Neanderthal and Denisovan population history (Slon et al., 2017; Zhang et al., 2020; Vernot et al., 2021; Zavala et al., 2021), focusing on the traces of mammal DNA, particularly of hominin origin. With caves often mitigating the full effects of the elements on organic materials such as bones, feces, and plant remains accumulated by humans, animals, and other natural agents, they provide a unique opportunity to study not only past human presence, but also past environments and human activities through DNA analyses.
Previous sedaDNA studies on cave sites in New Zealand, Australia, and North America demonstrate the potential to recover and identify plant DNA and reconstruct plant assemblages from >20,000 years ago (Willerslev, 2003; Haile et al., 2007; Haouchar et al., 2014; Seersholm et al., 2020). Moreover, a recent study at Solkota Cave in Georgia successfully retrieved up to 84,000-year-old mammal and plant DNA from stalagmites (Stahlschmidt et al., 2019), attesting to the unique conditions of preservation in caves.
The Armenian Highlands together with the southern Caucasus lie along an important route for early humans as they migrated out of Africa, with the earliest evidence dated to approximately 1.8 Ma at the Early Pleistocene site of Dmanisi in Georgia (Gabunia and Vekua, 1995; Egeland et al., 2010; Ferring et al., 2011; Gasparyan and Glauberman, 2022). It is an extremely diverse region, both topographically and climatically, and supports a variety of plant habitats from deserts, semi-deserts, and steppes to open woodlands, forests, and alpine meadows (Nakhutsrishvili, 2012; Fayvush and Aleksanyan, 2016). In Armenia, these vegetation units mainly follow altitudinal gradients of temperature and precipitation (Gulisashvili et al., 1975; Moreno-Sanchez and Sayadyan, 2005; Fayvush et al., 2013, 2017). The lower areas (480–1200 m) are characterized by semi-deserts, whereas a variety of steppic and forest vegetation covers the middle and upper mountain belts (1200–2000 m), and meadows and carpets characterize the subalpine and alpine areas (2200–4000 m; Fayvush et al., 2013, 2017). Only a few studies have described the Pleistocene paleovegetation in the Armenian Highlands (i.e., Bruch and Gabrielyan, 2002; Gabrielyan and Gasparyan, 2003; Joannin et al., 2010; Pinhasi et al., 2011), and information on Paleolithic plant use is even scarcer. Based on palynological evidence, the current vegetation types appear to have persisted throughout the Pleistocene with phases of expanded forest-steppe during more humid periods in response to climate oscillations (Joannin et al., 2010; Ollivier et al., 2010). Analysis of leaf imprints from several edible plants found in the area around Sisian, southern Armenia, suggests the environment was suitable for human life in the Plio-Pleistocene (Gabrielyan and Gasparyan, 2003; Fig. 1). Yet no indications of human presence have been found and, by extension, no evidence of human use of these plants for food. However, Acheulean localities recorded in the Vorotan Depression, north from Sisian, indicate human presence in this region as early as the Middle Pleistocene (Lyubin and Belyaeva, 2013). More direct evidence of human plant use comes from dyed and spun flax fibers from Upper Paleolithic strata at Dzudzuana Cave in Georgia, indicating that prehistoric hunter-gatherers in the southern Caucasus region were making cords, weaving baskets, or sewing garments (Kvavadze et al., 2009).

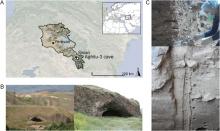
Figure 1. Aghitu-3 Cave. A) Map showing the location of the cave in the Republic of Armenia. B) View of the Aghitu basalt massif looking west into the valley of the Vorotan River (left) and a closer view looking west into the cave (right). C) SedaDNA sampling in between the basalt plates of AH III (top) and a section of the sediment profile in AH VI in the excavation pit (bottom). Photos by A. Kandel.
Here, we assess the potential of sedaDNA metabarcoding for reconstructing prehistoric plant assemblages from cave sediments by applying these methods to Aghitu-3 Cave in the Armenian Highlands. Specifically, we evaluate: 1) the potential of sedaDNA metabarcoding for recovering prehistoric plant assemblages from cave sediments; 2) whether prehistoric humans may have accumulated the identified plant remains; and 3) modern uses of the recovered plants to infer putative prehistoric human plant use. To this end, we combine plant sedaDNA metabarcoding with previous pollen evidence from Aghitu-3 Cave in southern Armenia, followed by analysis of modern uses for the recovered plants. Aghitu-3 Cave represents one of a handful of stratified Upper Paleolithic (Late Pleistocene) sites in the Republic of Armenia and contains a 15,000-yearlong record (from ∼39,000 to 24,000 cal BP) of Upper Paleolithic human settlement (Gasparyan et al., 2014). Plant remains identified throughout the sequence of Aghitu-3 include pollen and nonpollen palynomorphs, such as spores, algal and fungal remains, wood, and other undifferentiated plant parts (Kandel et al., 2017), implying good preservation of organic materials. The occupation horizons at Aghitu-3 yielded many finds, including stone artifacts, combustion features, and faunal remains, separated by archaeologically sterile layers with few finds (Kandel et al., 2017; Bertacchi et al., 2021). The observed interstratification creates an opportunity to test whether humans and animals accumulated the identified plant remains. Through the analysis of modern uses of the recovered plant taxa, we provide insight into their possible prehistoric use at Aghitu-3.
2. Materials and methods
2.1. Study site
Aghitu-3 Cave (39.5138° N, 46.0822° E, 1601 m a.s.l.) is situated 115 m above the Vorotan River in the Syunik Province of southern Armenia (Fig. 1A). With dimensions of 11 m depth, 18 m width, and 6 m height, it is the largest of several caves along the base of a basalt massif, the source of which was an eruption from nearby Mount Bughdablur between 126,000 and 111,000 years ago (Ollivier et al., 2010). The Vorotan River has cut through this basalt, forming a river valley and facilitating the movement of wildlife as well as people (Kandel et al., 2014). The current vegetation in this region is classified as ‘Pre- and Transcaucasian Stipa-Steppes’ according to the European Vegetation Map (Bohn et al., 2004). It is characterized by steppes and mountain meadows, with occasional small patches of oak forests (with Quercus macranthera) only on north-facing slopes with a more humid, favorable microclimate (Kandel et al., 2017). Archaeological excavations at Aghitu-3 started in 2009 and yielded many finds, including stone artifacts, faunal remains, bone tools, shell beads, charcoal, and pollen (Kandel et al., 2014, 2017; Bertacchi et al., 2021). Twelve geological and seven archaeological horizons (AHs) were originally identified. As the two uppermost horizons are mixed with Holocene debris, we focus on the intact Pleistocene stratigraphy of AHs VII–III, dated to ∼39,000–24,000 cal BP (Kandel et al., 2017).
The relatively dry cave environment of Aghitu-3, with sediments composed mainly of silt, clay minerals, volcanic ash, and basalt debris from the cave ceiling, has preserved many organic remains (Kandel et al., 2014, 2017; Bertacchi et al., 2021). Most of the sedimentary sequence consists of clayey silt, with somewhat more clay in AH VI compared to other layers; only in AH VII does the sediment grade to silty sand with depth (Kandel et al., 2014).
Remains obtained from palynological analyses were observed throughout the sedimentary sequence. Other organic remains found in the cave sediments include charcoal and faunal remains of birds, fish, micromammals, and macrofauna (Kandel et al., 2017; Bertacchi et al., 2021). The abundance patterns of these remains along the archaeological horizons indicate an association with human occupation. More organic remains were found in archaeological horizons that also contain high densities of lithics (AH VII, VI, and III), as compared to those considered almost sterile (containing <0.1% of the total number of lithics i.e., AH V and IV). Macrofaunal remains are rare in AH V and IV, followed by AH VII. A high abundance of mainly goats and horses was observed in AH VI and III (Kandel et al., 2017; Bertacchi et al., 2021). Anthropogenic modifications, including cut marks, percussion marks, and burn marks, were observed on several bones from these layers, and in AH VI, there was evidence that some of the fauna was accumulated by carnivores (Bertacchi et al., 2021).
The pattern of macrofaunal abundance matches that of the lithics as well as the microcharcoal concentrations, and together with the rare presence of carnivore remains and lack of coprolites suggest human occupation in AH VII, VI, and III, when the cave may have been used as a shelter for short-term stays for hunting (Taller et al., 2018). In contrast, the faunal remains from small mammals (lagomorphs and rodents) showed high rates of deposition, especially in AH V and IV, which have been associated with a reduced presence of humans in the cave (Kandel et al., 2017).
The abundance of organic remains throughout the sedimentary sequence suggests conditions of good preservation, while the stratification of human occupation layers (AHs VII, VI, and III) interspersed with archaeologically sterile layers (AH V and IV) creates the opportunity to make inferences about possible human plant use at Aghitu-3 Cave during the Upper Paleolithic.
2.2. Sediment sampling for sedimentary ancient DNA
The Aghitu-3 excavation profile (>5.5 m) consists mainly of dry silt, which is dominant throughout and interspersed with basalt debris from the cave ceiling. We collected sedaDNA samples in September 2019 over the full depth of the profile. As AH III contains high amounts of platy basalt (Fig. 1C), samples were taken from accessible sediments in between the basalts. In the lower horizons (AH VII and VI), which contain much less basalt, we did not sample close to boulders and avoided including small rocks.
The cave sedimentary sequence was sampled while wearing personal protective equipment (hairnets, facemasks, double gloves) and using sterile instruments. Sampling tubes and scalpel holders were UV irradiated in preparation for the sampling, and scalpel holders were cleaned with bleach in between each sample to avoid cross-contamination of DNA. First, 1–2 cm of surface material was removed. Then two consecutive cuts were made with sterile scalpels before sample collection in a sample tube, following the procedure described by Epp et al. (2019). Samples were kept cool during transport to the ancient DNA facilities at the University of Oslo, where they remained at −20 °C until DNA extraction.
2.3. DNA analyses
DNA extraction and amplification DNA was extracted from 25 samples and three negative controls at the dedicated ancient DNA laboratory, Department of Biosciences, University of Oslo. We extracted DNA from 90 to 190 mg of sediment using the sediment extraction protocol as described by Rohland et al. (2018) with silica spin columns and binding buffer D. We increased the elution buffer volume to 100 μL. See Supplementary Online Materials (SOM) S1 for further details.
Plant chloroplast DNA was amplified using the trnL-gh primers that target the P6 loop from the trnL-intron (Taberlet et al., 2007). These primers are specifically designed for amplification of degraded plant DNA and are widely used in plant sedaDNA studies. Both primers were tagged with a unique 8 or 9 base pair long barcode at the 5′ end to allow for multiplexing as described by Voldstad et al. (2020). We conducted six polymerase chain reaction (PCR) replicates per sample and extraction negative control, and included 30 PCR negative controls. Amplifications of DNA were carried out in a final volume of 25 μL containing 5 μL of DNA extract and 0.2 μM of each primer. The amplification mixture further contained 2U of AmpliTaq Gold DNA Polymerase with Buffer II (Applied Biosystems), 1 × Buffer II, 2.5 mM MgCl2, 0.2 mM of each dNTP, and 0.32 μg/μL of bovine serum albumin (BSA, Roche Diagnostic). The mixture was denatured at 95 °C for 10 min, followed by 45 cycles of 30 s at 95 °C, 30 s at 50 °C, 1 min at 72 °C, and a 10-min final elongation at 72 °C.
Polymerase chain reaction products were visualized with agarose gel electrophoresis. Equal volumes of PCR products with similar band intensity were mixed and purified using the MinElute Purification kit (Qiagen GmbH, Hilden, Germany), including PCR products that did not show a band. Concentrations of the pools with purified PCR products were measured on a Qubit 2.0 with the Qubit dsDNA HS kit (ThermoFisher Scientific), and pools were subsequently combined in an equimolar superpool. Libraries were built from the resulting superpool using the KAPA HyperPrep DNA kit (Roche) and sequenced on the Illumina HiSeq 4000 at the Norwegian Sequencing Centre.
DNA sequence analyses and filtering Initial sequence data processing was done using the OBITools package v. 1.2.12 (http://metabarcoding.org/obitools/doc/index.html; Boyer et al., 2016). Forward and corresponding reverse reads were assembled using ‘illuminapairedend,’ followed by sample assignment with ‘ngsfilter.’ We removed reads with a quality score <40, <100% tag match, >3 mismatches with the primers, shorter lengths than expected (<8 bp), and those containing ambiguous nucleotides. Unique reads were merged and singletons removed. Amplification and sequencing errors were identified using ‘obiclean,’ with a threshold ratio of 5% for reclassification of sequences identified as ‘internal’ to their corresponding ‘head’ sequence. Finally, sequences were compared to a taxonomic reference library using ‘ecotag.’ The reference library was prepared using the global EMBL sequence database (release 142; January 2020; http://www.ebi.ac.uk/embl/) and the NCBI Taxonomy database (https://www.ncbi.nlm.nih.gov/taxonomy) by performing an in-silico PCR with the ecoPCR software v. 1.0.1 (Ficetola et al., 2010).
To minimize the chance of misidentification, we filtered the identified sequences in R v. 4.1.0 (R Core Team, 2021) to further remove: 1) sequences occurring in negative controls, 2) sequences with a percentage identity <97.5%, 3) read counts <10 reads per sequence in a PCR replicate, and 4) PCR replicates with <10 reads. As the used reference database contains limited data from the Caucasus region, it is possible that DNA sequences were misassigned to a sister taxon, or to a more narrow taxon than appropriate, causing potential overconfidence in the taxonomic assignments. To mitigate these potential biases, a taxonomic expert checked the taxonomic annotations to assess the likeliness of occurrence in the region at the time. Based on this assessment, several taxonomic assignments were adjusted (see SOM Table S1) and additional filtering steps were performed removing sequences occurring in a single PCR replicate over the entire data set, sequences with a percentage identity <98%, and sequences without family-annotation or that were deemed unlikely for the period and location. Samples consisting of <2 positive replicates after filtering were considered empty and their read counts were set to 0. Remaining unique DNA sequences were designated as molecular operational taxonomic units (MOTUs). An overview of the filtering steps and their effect on the size of the data set, as well as a list of manually removed taxa, can be found in SOM Table S2.
The filtered sequence data were corrected for the amount of input material (SOM Table S3). Polymerase chain reaction replicates were merged while calculating the total read counts for each sample as well as the number of positive replicates. The total read counts per sample were subsequently log-transformed to correct for the exponential increase in read counts during PCR amplification.
All raw read data of this study are available at the European Nucleotide Archive under study accession number PRJEB54625. Scripts for read data processing are available at https://github.com/terschure/dataprocessing_AGH. All processed data are available in SOM Tables S1 and S4–S6.
2.4. Pollen data
Sediment samples for palynological analysis (including pollen) were obtained along the sediment profile walls during excavations in 2011 and 2013 (Kandel et al., 2017). Per sample, 50 g of sediment was processed using 30% HCl and 30% HF to remove carbonates and silica. Tablets of spores (Lycopodium) were added to each sample to calculate pollen concentrations (Stockmarr, 1971). Sodium polytungstate (SPT) was used to separate particle fractions by gravity. The samples were sieved with a 6 μm filter and centrifuged with SPT to separate particle fractions. Two strew-mounted slides were prepared from each residue with glycerin gelatin as a mounting medium. The resulting palynological data have previously been used for qualitative analysis of the vegetation and environmental conditions at Aghitu-3 (Kandel et al., 2017). In this study, we focus on the pollen data.
2.5. Plant-use assessment
For the assessment of plant use by humans, we selected all taxa from the pollen and sedaDNA records that were identified to genus or species. From this list, we limited our analyses to those species that are included in the Flora of Armenia (Takhtajan, 1954–2009). For taxa determined at genus level (DNA and pollen) or as pollen types, both of which are represented by several species, we used only Armenian species for the compilation of information. For example, of 472 accepted species of Artemisia, 23 occur in Armenia, and only these were assessed for evaluating the potential plant use of the genus Artemisia in this study. The taxonomic status of these Armenian species was checked with the Plants of the World Online database (http://www.plantsoftheworldonline.org/) for their validity and potential synonyms.
Potential uses of plants were identified using the PlantBITES database (Altolaguirre et al., 2021, unpublished data) from “The Role of Culture in Early Expansions of Humans” (ROCEEH) project, the “Plants For A Future” database (https://pfaf.org; Fern, 1997), the “Useful Temperate Plants” database (http://temperate.theferns.info/; Fern, 2019), as well as botanical and ethnobotanical publications (e.g., Rivera et al., 2012; Fleischhauer et al., 2016). In cases where, after thorough research, no information was available, we considered the taxon as not having any uses as known thus far (see Table 1). We evaluated the following categories of use: edible, medicinal, dye, repellent, and other (e.g., string making, thatching, weaving, strewing; i.e., the scattering of fragrant or insect repellent plants). Plants of the ‘edible’ category were further divided into whether parts of the respective plant can be eaten in large amounts, i.e., served as bulk food, or if the plant is used as a flavoring (condiment, herb, spice, tea). For each taxon, we scored its use per category as to whether it is useful or not as determined by being reported in the literature. For taxa identified to genus level and comprising several species, we scored the genus as being useful if one or several of the Armenian species listed could be classified as useful. In cases where all Armenian species of a genus are considered useful, this was noted in the table. If none of the Armenian species of this genus had any reported use, the genus was considered as not having any uses as known thus far.
Table 1. List of Armenian plant genera and species documented by sedaDNA and pollen from Aghitu-3 Cave and their assignment to plant-use categories based on information in the ROCEEH project database PlantBITES (Altolaguirre et al., 2021, unpublished data), the Plants For A Future database (https://pfaf.org; Fern, 1997), the Useful Temperate Plants database (http://temperate.theferns.info/; Fern, 2019), as well as botanical and ethnobotanical publications (e.g., Rivera et al., 2012; Fleischhauer et al., 2016). The number of species in Armenia refers to those listed in the Flora of Armenia (Takhtajan, 1954–2009). The taxonomic status was checked with the Plants of the World Online database (http://www.plantsoftheworldonline.org/) and the number of accepted species according to this database is given in brackets.
| Assigned Armenian taxa | n species in Armenia | Proxy | Bulk food | Flavoring | Medicinal | Dye | Repellent | Other uses | Other uses being: |
|---|---|---|---|---|---|---|---|---|---|
| Forbs | |||||||||
| Acoraceae | |||||||||
| Acorus calamus L. | Pollen | – | √ | √ | – | √ | √ | Basketry, strewing, thatching, weaving | |
| Amaranthaceae | |||||||||
| Atriplex sp. | 12(11) | DNA | √ | – | √ | – | – | – | |
| Beta vulgaris type | 5 | Pollen | √ | – | √ | – | – | – | |
| Chenopodium sp. | 12(11) | DNA | √a | – | √ | √ | √ | – | |
| Asteraceae | |||||||||
| Achillea pseudoaleppica Hausskn. ex Hub.-Mor. | DNA | –b | –b | √ | –b | –b | –b | ||
| Artemisia sp. | 24(23) | Both | – | √ | √ | – | √ | √ | Strewing |
| Centaurea sp. | 88 | Pollen | – | – | – | √ | – | – | |
| Cirsium sp. | 22(11) | DNA | √ | – | √ | – | – | √ | Tinder |
| Matricaria matricarioides (Less.) Porter ex Britt. | Pollen | – | √ | √ | – | √ | – | ||
| Boraginaceae | |||||||||
| Lappula sp. | 8(6) | DNA | –b | –b | –b | –b | –b | –b | |
| Lithospermum officinale L. | DNA | – | √ | √ | √ | – | – | ||
| Brassicaceae | |||||||||
| Alyssum sp. | 24(15) | DNA | – | – | –b | –b | –b | –b | |
| Brassica nigra (L.) W.D.J.Koch | DNA | √ | – | √ | – | √ | – | ||
| Brassica oleracea L. | DNA | √ | – | √ | – | √ | – | ||
| Isatis sp. | 13(12) | DNA | √ | √ | √ | √ | – | √ | Preservative |
| Lepidium sp. | 12(12) | DNA | – | √ | √ | –b | –b | –b | |
| Raphanus raphanistrum L. | DNA | √ | – | √ | – | – | – | ||
| Sterigmostemum sp. | 2(2) | DNA | –b | –b | –b | –b | –b | –b | |
| Caprifoliaceae | |||||||||
| Knautia arvensis type | 2(2) | Pollen | – | √ | √ | – | – | – | |
| Convolvulaceae | |||||||||
| Convolvulus arvensis type | 2 | Pollen | – | √ | √ | √ | – | √ | String |
| Ephedraceae | |||||||||
| Ephedra distachya type | 1 | Pollen | – | √ | √ | – | – | – | |
| Fabaceae | |||||||||
| Astragalus sp. | 144 | DNA | – | √ | √ | – | – | – | |
| Hedysarum sp. | 8 | DNA | –d | –d | – | – | – | – | |
| Onobrychis sp. | 25(23) | DNA | –c | √c | –c | –c | –c | –c | |
| Pisum sativum L. | DNA | √ | – | √ | – | – | – | ||
| Malvaceae | |||||||||
| Malva neglecta type | 3 | Pollen | √ | – | √ | √ | – | √ | Toothbrush |
| Polemoniaceae | |||||||||
| Polemonium caeruleum L. | Pollen | – | – | √ | – | – | – | ||
| Polygonaceae | |||||||||
| Rumex scutatus L. | DNA | √ | – | √ | √ | – | – | ||
| Ranunculaceae | |||||||||
| Thalictrum sp. | 6 | Pollen | – | √ | √ | – | – | – | |
| Typhaceae | |||||||||
| Typha sp. | 10 | Pollen | √ | – | √ | – | – | √ | Tinder, thatching, weaving, stuffing |
| Viscaceae | |||||||||
| Viscum album L. | Pollen | – | √ | √ | – | – | – | ||
| Graminoids | |||||||||
| Poaceae | |||||||||
| Alopecurus myosuroides Huds. | DNA | –b | –b | –b | –b | –b | –b | ||
| Phragmites australis (Cav.) Trin. ex Steud. | Pollen | √ | – | √ | √ | – | √ | Thatching, string, weaving | |
| Trees and shrubs | |||||||||
| Betulaceae | |||||||||
| Betula sp. | 3(2) | Pollen | – | √a | √ | – | – | √ | Drinking vessels |
| Carpinus sp. | 3 | Pollen | – | – | √ | √ | – | – | |
| Corylus sp. | 2 | Pollen | √ | – | √ | – | – | √ | Basketry, tannin |
| Buxaceae | |||||||||
| Buxus sempervirens L. | Pollen | – | – | √ | – | – | – | ||
| Cannabaceae | |||||||||
| Celtis sp. | 2 | Pollen | √a | – | –b | – | – | – | |
| Fagaceae | |||||||||
| Quercus sp. | 6(4) | DNA | – | √ | √ | √ | √ | √ | Gum, basketry, tannin |
| Quercus frainetto type | Pollen | – | √ | √ | √ | √ | √ | Gum, basketry, tannin | |
| Quercus coccifera type | Pollen | – | √ | √ | √ | √ | – | ||
| Juglandaceae | |||||||||
| Juglans regia L. | Pollen | √ | – | –b | – | – | – | ||
| Pinaceae | |||||||||
| Pinus sp. | 6(4) | Pollen | – | √a | √ | √ | – | √ | String, tannin, pitch, turpentine |
| Salicaceae | |||||||||
| Salix sp. | 14 | Pollen | √ | – | √ | √ | √ | √ | String, resin, adhesive, basketry, stuffing, tannin |
| Sapindaceae | |||||||||
| Acer sp. | 7 | Pollen | √ | – | √ | √ | – | – | |
- a
-
True for all Armenian species.
- b
-
No information found, designated as not useful.
- c
-
No information on Armenian species, information on Onobrychis viciifolia was used.
- d
-
Some Chinese and American species are reported to be edible, but those known to occur in Armenia are not edible.
2.6. Statistical analyses
All statistical analyses were performed in R v. 4.1.0 (R Core Team, 2021). To visualize changes in abundance of plant taxa over time, we created stratigraphic plots with the ‘rioja’ package v. 0.9-26 (Juggins and Juggins, 2020; SOM Figs. S1–S3).
To investigate plant assemblages, we used nonmetric multidimensional scaling (NMDS) based on Bray–Curtis dissimilarities, allowing the visualization of dissimilarities in composition across samples. A stress level of <0.2 for NMDS is considered acceptable for this purpose (Clarke, 1993). We grouped samples according to their archaeological horizons and tested for differences between groups by performing a permutational multivariate analysis of variance (perMANOVA) with 999 permutations using the adonis function in the ‘vegan’ package v. 2.5-7 (Oksanen et al., 2020). For pairwise perMANOVAs, we corrected p-values for multiple testing using the Bonferroni method (Bland and Altman, 1995) of the p.adjust function. We tested the perMANOVA assumption of homogeneity of multivariate dispersions using the permutest function with 999 permutations.
To facilitate comparisons between sedaDNA and pollen diversity and relate these to plant-use information, we reduced the sedaDNA data set from MOTUs to species and genera, summing the different MOTUs of the same species or genus together. This reduced data set was used to determine the taxonomic richness and the inverse Simpson index using Hill numbers (q = 0 and q = 2) with the hill_taxa function of the ‘HillR’ package v. 0.5.1 (Li, 2018). We used the nonparametric Mann–Whitney–Wilcoxon test to identify differences in plant diversity among archaeological horizons, available under the function wilcox_test from the ‘rstatix’ package v. 0.7.0 (Kassambara, 2021) with standard settings, including a Holm correction of p-values (Holm, 1979). We applied a significance level of α = 0.05 for all statistical tests.
3. Results
3.1. Brief description of the data
We obtained 168.8 million raw DNA sequence reads. After filtering, the DNA data set consisted of 3.3 million reads from 21 samples, leaving four samples with no sequences (SOM Table S1). Read counts per sample, corrected for the amount of input material, averaged 305 ± 38 log-transformed reads per mg of sediment (excluding empty samples; Fig. 2A; SOM Table S4). The number of positive replicates per sample averaged 4.3 ± 0.7 (excluding empty samples; Fig. 2A; SOM Table S5). The filtered DNA data set contained 128 unique MOTUs, corresponding to 10 plant families, 20 genera, and six species (see SOM Figs. S1 and S2 for detailed stratigraphic plots). Forbs (nongraminoid herbaceous plants) account for 116 MOTUs and more than 93% of the log-transformed read abundance, followed by graminoids, with 11 MOTUs at approximately 6% and trees with 1 MOTU at <0.5%.

Figure 2. Plant diversity per archaeological horizon (AH) based on sedaDNA and pollen analysis. A) The total concentrations of plant sedaDNA (brown tones) and pollen (green tones) per sample and the inverse Simpson index of the recovered plant genera and species with respect to depth. The size of the circles indicates for sedaDNA, the number of positive replicates kept after filtering, and for pollen, the total pollen sum. The schematic profile drawing provides an overview of the sediment profile; however, the thickness and depth of the indicated layers vary spatially across the excavated area (Kandel et al., 2017). B) Boxplots of the total number of sedaDNA molecular operational taxonomic units (MOTUs; left: brown tones) and pollen taxa (right, green tones) detected per layer. Center line, median; box limits, upper and lower quartiles; whiskers, extremes of the data. A total of 25 sedaDNA samples (AH III, n = 5; IV, n = 2; V, n = 2; VI, n = 10; VII, n = 6) and 38 pollen samples (AH III, n = 4; IV, n = 8; V, n = 3; VI, n = 13; VII, n = 10) were compared. The coloring of the circles and bars indicates the AH, with darker colors for deeper horizons. Shaded areas on the right side of the figures indicate the relative intensity of human occupation (very low, low, medium, and high) as reported by Kandel et al. (2017). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article).
Twenty-seven of the 38 previously analyzed palynological samples contained pollen. Altogether, 39 taxa were identified, covering 28 families, 24 plant genera, and seven species, all corresponding to taxa occurring in Armenia. Total pollen concentrations averaged to 278 ± 75 grains per gram of sediment (excluding empty samples; Fig. 2A). Forbs were the most abundant plant group with 26 pollen types accounting for more than 70% of the total concentration. Trees, represented by 12 pollen types, accounted for 17.5%, followed by graminoids at 12%. Detailed pollen concentration diagrams can be found in SOM Figure S3 and the corresponding data in SOM Table S6.
3.2. Recovery of plant taxa
Comparison of pollen and sedaDNA Across pollen and sedaDNA data sets, a total of 32 plant families, 43 genera, and 14 species were identified, with an overlap between the two data sets of six plant families (19%), two genera (5%), and no overlap in detected species. Regarding the plant families, 22 are unique to the pollen data set, including Betulaceae and Pinaceae, and four are unique to the sedaDNA data set (Boraginaceae, Fabaceae, Lamiaceae, and Polygonaceae). The six plant families occurring in both data sets are Asteraceae, Brassicaceae, Amaranthaceae, Fagaceae, Plumbaginaceae, and Poaceae. Asteraceae (55%) represents the most abundant plant family in the pollen record, whereas Brassicaceae (44%) dominates the sedaDNA data set, followed by Fabaceae (21%). The plant genera detected in both data sets are Artemisia (wormwoods) and Quercus (oaks). Another 23 genera were uniquely detected by pollen compared to 18 by sedaDNA analysis.
Plant assemblages across archaeological horizons Among the analyzed samples, 21 sedaDNA and 27 pollen samples contained identifiable plant taxa and these were used to investigate the plant assemblages. The number of samples per horizon varied between two and 10 (Fig. 2). No sedaDNA was recovered from the two samples taken from AH IV. Nonmetric multidimensional scaling showed visual separation of AH III from all other AHs for both pollen and sedaDNA data sets, and sedaDNA samples in particular cluster more closely for some horizons (AH VII and VI) than for others (AH V; Fig. 3). Permutational multivariate analysis of variance on Bray–Curtis dissimilarities derived from both data sets (sedaDNA and pollen) indicated significant differences between plant assemblages among AHs (sedaDNA adonis F3,20 = 2.53, r2 = 0.31, p < 0.001; pollen adonis: F4,26 = 1.70, r2 = 0.24, p = 0.011). The assumption of homogeneity of dispersion among AHs was supported by nonsignificant permutest results (sedaDNA: p = 0.158, pollen: p = 0.352). Post hoc pairwise perMANOVAs on subsets of the sedaDNA data indicated statistically significant (Bonferroni-adjusted p < 0.05) differences in plant assemblages between AH III and VI, between AH III and VII, and between sets of AHs, VII-VI and V-IV, and VII-VI and III. The same analysis for the pollen data set revealed a significant difference between the AH VII-VI set and III, but no differences in plant assemblages were found using pairwise comparisons of individual AHs based on Bonferroni-adjusted p-values.

Figure 3. SedaDNA (left; brown tones) and pollen (right; green tones) nonmetric multidimensional scaling (NMDS) ordination of Bray–Curtis dissimilarity of samples (small circles) and averages per archaeological horizon (big circles). The stress levels of 0.138 and 0.134 are under the cut-off value of 0.2 as posed by Clarke (1993) to indicate an interpretable ordination. The coloring is indicative of the archaeological horizon (AH), with darker colors for deeper horizons. One pollen sample outlier (sample 11 from AH VI, depth = −2.26, NMDS1 = −4.06, NMDS2 = −0.31, containing only Juglans sp. pollen) was excluded from visualization here, but included in the statistical analysis. Archaeological horizon IV is missing from the sedaDNA NMDS (left) as associated samples were considered empty. Plant assemblages among AHs were significantly different (p < 0.05) for both sedaDNA and pollen. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article).
Plant richness was highest in the lower archaeological horizons, with AH VII having a median of 31 MOTUs (interquartile range [IQR] = 19.75–42.75, n = 6) and three pollen types (IQR = 0.5–5.5, n = 10) and AH VI having a median of 38 MOTUs (IQR = 28.5–40.75, n = 10) and three pollen types (IQR = 1–6, n = 13; Fig. 2A).
In AH VII, the pollen record was dominated by Asteraceae, whereas Brassicaceae dominated the sedaDNA record with five genera (Brassica sp.: cabbage, turnip, mustard; Isatis sp.: woad; Lepidium sp.: cress; Raphanus sp.: radish; and Sterigmostemum sp.). Both records indicate the presence of these families, as well as Amaranthaceae, identified by sedaDNA as mainly Atriplex sp. (orach) and Chenopodium sp. (goosefoot). Other plant families in this AH detected by both records are Poaceae (Alopecurus sp.: foxtail grass, and other unidentified grasses) and Fagaceae (i.e., Quercus sp.: oak). Specific to the pollen record in AH VII were traces of other trees (Pinus sp.: pine; Acer sp.: maple), some forbs and graminoids (Apiaceae, Caryophyllaceae, Cyperaceae, Convolvulus sp.: bindweed), and the gymnosperm shrub Ephedra sp., whereas the sedaDNA record counted another 10 genera.
In AH VI, one pollen sample (sample 41, depth = −4.26 m) stands out with higher pollen counts (n = 218; Fig. 2A) and higher diversity than any other analyzed pollen samples, containing 28 of the 38 identified pollen types detected throughout the sequence. Particularly Phragmites sp. and Asteraceae pollen were abundant in this sample. Asteraceae remained present in the pollen record throughout AH VI, with concentrations increasing with depth. In the sedaDNA record, Brassicaceae and Amaranthaceae remained abundant, whereas Fabaceae (particularly Astragalus sp.: milkvetch) showed a much higher presence than in AH VII.
Moving up the sequence from AH VI to AH V, the median number of plant taxa declined to 3.5 MOTUs (IQR = 2.75–4.25, n = 2) or 2 pollen taxa (IQR = 1–3.5, n = 3). Detected plant taxa in AH V included Asteraceae and Poaceae in both sedaDNA and pollen, Fabaceae and Lamiaceae only in the sedaDNA record, and Amaranthaceae, Juglans regia (walnut), Quercus sp., and Pinus sp. only in the pollen.
In AH IV, sedaDNA detected no plant MOTUs and the median number of pollen taxa was 1.5 (IQR = 0–3.25, n = 8). Asteraceae pollen concentrations increased compared to AH V and pine pollen was continuously detected. Other pollen finds included Ericaceae, Amaranthaceae, Brassicaceae, and low concentration of J. regia at the bottom of this AH.
In AH III, the median number of MOTUs increased to 12 (IQR = 9–14, n = 5), whereas pollen taxa further decreased to a median of 0.5 (IQR = 0–1.5, n = 4). A high concentration of Artemisia sp. pollen was found, as well as Pinus sp. and Amaranthaceae. In the sedaDNA data, this AH was dominated by Fabaceae, particularly Astragalus sp., but also some Pisum sp. (pea). Further finds included several Brassicaceae (Brassica nigra: black mustard; B. oleracea: wild cabbage and cultivars; and Raphanus raphanistrum) and one taxon (Pisum sp.) from the legumes family (Fabaceae).
No significant differences in plant richness between individual AHs were found for either sedaDNA or pollen. Grouping AHs together provided more statistical power and revealed a significant decrease in sedaDNA-based plant richness from the AH VII-VI set to AH V-IV (Mann–Whitney U = 3, p = 0.021), followed by a significant increase toward AH III (Mann–Whitney U = 7.5, p = 0.021; see SOM Table S7). This same grouping did not result in significant differences in pollen-based plant richness.
3.3. Potential use of recovered plant taxa
Useful plant data The potential use of the detected plant taxa was determined for the plants identified to genus and species level, reducing the sedaDNA data set to 21 taxa, and the pollen data set to 25 pollen types. With the focus on Armenian species, most of these plants were considered useful in at least one, and many of them in multiple use categories (i.e., bulk food, flavoring, medicinal, dye, repellent, other), including 16 of the taxa identified with sedaDNA, and 20 of the taxa identified with pollen analysis. Those considered not useful are as follows: Alopecurus myosuroides, Lappula sp. (stickseed), Alyssum sp. (alyssum), Sterigmostemum sp., and Hedysarum sp. (sweetvetch; Table 1).
Edible plants A total of 15 edible plant taxa were detected by sedaDNA analysis, nine of which categorized as bulk foods, and seven usable as flavoring. Pollen analysis detected 18 edible plant taxa: six bulk food and 11 flavoring (Table 1; Fig. 4). The bulk food taxa detected by sedaDNA were primarily found in AH VII, VI, and III and included several members of the Brassicaceae (e.g., B. nigra, B. oleracea, R. raphanistrum, Isatis sp.) and Amaranthaceae (i.e., Atriplex sp. and Chenopodium sp.) as well as Rumex scutatus (French sorrel), Cirsium sp. (plume thistle), and Pisum sp. The pollen record indicates a high richness in bulk food in one particular sample (depth = −4.26 m) including Beta vulgaris (beet) type pollen and those of Phragmites sp. (reed) and Typha sp. (bulrush), whereas other pollen samples include one bulk food taxon at most, from either Celtis sp. (nettle tree), Juglans sp., or Acer sp. Plant taxa that could be used for flavoring were more consistently present in the pollen record, and were also detected in many sedaDNA samples throughout the sedimentary sequence (Fig. 4).

Figure 4. Useful plants per archaeological horizon (AH) based on the reduced sedaDNA and pollen datasets to genus- and species-level identifications. A) The number of plant taxa considered useful per category and proxy (sedaDNA, pollen, and both proxies combined) of the total number of genus- and species-level identifications. B) The number of useful plant taxa per category over depth for sedaDNA (left; brown tones) and pollen (right; green tones). C) The number of useful and not-useful plant taxa per category, summarized per AH for sedaDNA (top) and pollen (bottom). The coloring of the bars is indicative of the AH, with darker colors for deeper horizons. Shaded areas on the right side of the figures indicate the relative intensity of human occupation (very low, low, medium, and high) as reported by Kandel et al. (2017). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article).
Other uses Fifteen of the 16 useful plant taxa detected with sedaDNA were categorized as medicinal compared to 21 of the 24 useful plant taxa detected through pollen (Table 1). Both records indicate consistent presence of these taxa throughout the sedimentary sequence (Fig. 4).
Plant taxa possibly used for dye were similarly detected throughout the pollen record but limited to AH VII and VI in the sedaDNA record. Those detected by sedaDNA included R. scutatus, Lithospermum sp. (gromwell), Isatis sp., and Chenopodium sp., while the pollen record distinguished eight different taxa, of which four trees (two Quercus types, Pinus sp., and Acer sp.), Pinus sp. in particular, being present throughout most of the sedimentary sequence.
The category of repellents encompassed five taxa detected with sedaDNA and three detected with pollen. These include several of the taxa also categorized as bulk foods and/or flavoring (Brassica sp., Artemisia sp., Chenopodium sp., and Quercus sp.), and sedaDNA from these categories showed similar patterns (Fig. 4). Repellent taxa found by pollen analysis were Acorus calamus (sweet flag), Artemisia sp., and Matricaria matricarioides (wild chamomile) type (Table 1).
Ten taxa were assigned to the category of ‘other uses,’ of which two were found with both proxies (Quercus sp. and Artemisia sp.), two others uniquely with sedaDNA (Cirsium sp. and Isatis sp.), and eight with pollen (Table 1; Fig. 4). Several of the pollen taxa belonging to this category are trees (Pinus sp., Quercus sp., and Betula sp.), while others are grasses (Phragmites sp.) and forbs (A. calamus, Artemisia sp., Convolvulus sp. and Typha sp.).
4. Discussion
We combined evidence of plant presence from sedaDNA metabarcoding and pollen data with information on known plant uses to assess the potential of sedaDNA metabarcoding for reconstructing prehistoric plant assemblages associated with human occupation. Pollen and sedaDNA analyses enabled us to reconstruct the plant assemblages, while the assessment of plant utility provided insight into potential human plant use at Aghitu-3 Cave during the Upper Paleolithic. In the following section, we discuss the effects of taphonomic processes on the recovery of plant taxa by pollen and sedaDNA analyses, the observed changes in plant assemblages over time, and the potential prehistoric human plant use at Aghitu-3.
4.1. Taphonomy and recovery of plant sedaDNA and pollen
Plant sedaDNA was recovered throughout the Aghitu-3 Cave sequence, consistent with the numerous organic finds throughout the stratigraphy as reported by Kandel et al. (2017). By contrast, the recovery of pollen appeared more variable, and pollen numbers were generally low (Fig. 2A). The pathways through which vegetation travels from outside the cave to the cave floor vary, including transportation by wind, by animals (for nests, burrows, on their bodies, or in fecal matter), or by humans (Hunt and Fiacconi, 2018). Alternatively, plant remains could come directly from vegetation growing at the cave entrance (Hunt and Fiacconi, 2018). As Aghitu-3 is a relatively wide and shallow cave, it is possible that plant remains were deposited in the cave without animal or human agency. Comparing the different botanical proxies and integrating these with other evidence from the site facilitates further inferences about the origin of the identified plant diversity.
Origin and taphonomy of plant sedaDNA and pollen Pollen and sedaDNA records are differentially affected by source productivity and taphonomic processes of dispersal, transfer, and deposition (Niemeyer et al., 2017; Alsos et al., 2018; Giguet-Covex et al., 2019), and both proxies signify different records. SedaDNA is generally considered of local origin (Jørgensen et al., 2012; Alsos et al., 2018), whereas pollen (especially from wind-pollinated plants, such as many trees and grasses) may originate from a wide area as it is distributed regionally through the air (Birks and Bjune, 2010; Parducci et al., 2018). Moreover, plant DNA in sediments can originate from most plant parts, but is unlikely to source from pollen due to the robustness of the pollen grain wall and the low amount of DNA it contains (Parducci et al., 2017; Sjögren et al., 2017; Clarke et al., 2020). Furthermore, the primers used in this study are unsuitable for analyzing DNA in pollen grains as they target chloroplast DNA, which is especially low or absent in pollen (Zhang et al., 2003; Ellis et al., 2008).
Postdepositional factors impacting pollen and sedaDNA could additionally produce differences in the recovery of plant data by the two proxies. Although pollen grains are highly robust, sedaDNA is known to degrade significantly as a result of temperature fluctuations, low pH, and high oxygen and water content, thereby decreasing the amount of detectable DNA over time (Willerslev and Cooper, 2005). DNA is better preserved in sediments with a high mineral content and at low temperatures. Minerals can bind to and protect DNA, whereas low temperatures thermally stabilize DNA against chemical degradation (Torti et al., 2015). Desiccated sediments can also prevent DNA damage and breakage by strongly decreasing the effects of hydrolysis. In addition to degradation, DNA may be translocated after deposition, either through the movement of sediment particles containing DNA, or the movement of DNA in solution, also known as DNA leaching (Haile et al., 2007; Pedersen et al., 2015). When there is a more or less constant input of plant remains through time and no DNA leaching, as is the case for many lake sediments, sedaDNA recovery is usually lower in older sediments due to degradation processes (Parducci et al., 2017). We do not observe such patterns of recovery; we recover higher DNA concentrations in the lower archaeological horizons (AH VII and VI) and overall DNA concentrations were higher compared to the younger horizons (AH V, IV, and the lower part of AH III) without obvious evidence of change in the sediment composition (Fig. 2A).
Aghitu-3 cave has been a dry and stable environment with a more or less similar sediment composition throughout the stratigraphy, with sediments composed mainly of silt, clay minerals, volcanic ash, and basalt debris from the cave ceiling (Kandel et al., 2014, 2017; Bertacchi et al., 2021). Preliminary micromorphological analyses do suggest some movement of sediment during specific horizons. Specifically, low-energy surface flow has been observed in AH V and below, whereas AH IV and III are characterized by erosion and eolian transport (Kandel et al., 2017). Some evidence for bioturbation was found as well as evidence for freeze-thaw processes in the form of lenticular microstructures overprinting pre-existing laminations (Kandel et al., 2017). Thawing could imply some degree of downward percolation of moisture as well as degradation of DNA through hydrolysis or changes in temperature, possibly decreasing sedaDNA concentrations. These processes suggest that, at least at a microscale level, DNA concentration may have been affected by postdepositional processes. To fully understand these processes and firmly establish the stability of plant DNA at Aghitu-3, DNA extraction of microscale sediment samples together with in-depth micromorphological analyses is needed, such as those recently presented for mammal DNA from Denisova cave (Massilani et al., 2022). Hereby, heterogeneity in taxonomic composition, not just among adjacent macroscale samples (as we observe) but also among microscale samples, would support limited movement of DNA through leaching. Nevertheless, the freeze-thaw and bioturbation processes observed in the preliminary micromorphological analyses at Aghitu-3 did not cause any major, visible disturbance affecting the distribution horizons of the original deposits. The current evidence of the homogeneity of the cave sediments together with the preliminary micromorphological analyses therefore suggest that postdepositional processes have not played a major role leading to differences in DNA concentrations between archaeological horizons at Aghitu-3, but that these differences are rather a result of the transfer of plant remains into the cave. Nevertheless, there is a need for explicit analyses on microstratigraphy and sedaDNA of plants for disentangling the role of taphonomy and human behavior in the plant profile that can be retrieved from these kinds of sites.
Source and transfer of recovered plant sedaDNA and pollen into Aghitu-3 The sedaDNA record at Aghitu-3 indicates relatively high plant DNA concentrations throughout AH VII and VI, whereas pollen concentrations are low in these horizons (Fig. 2A). In AH III, where most archaeological remains were found, both sedaDNA and pollen abundance increased relative to the sterile underlying layers. However, the increase in the pollen record is driven by a peak in abundance in one sample, and a higher peak was observed in AH IV, whereas for the sedaDNA, the increase is consistent across all samples taken from AH III (Fig. 2A). The Asteraceae family dominates the pollen record and the observed peaks, and although several known wind-pollinated taxa were found (including grasses, oak, and pine), their relatively low concentrations compared to primarily insect-pollinated Asteraceae suggest wind-transportation of plant remains into the cave was limited (SOM Figs. S1 and S2). The combination of low pollen concentrations with high sedaDNA abundance suggests animals or humans may have actively brought plant remains into the cave during AH VII, VI, and AH III.
It can be expected that plants are more likely to establish inside the cave or its entrance during periods of reduced occupation by either animals or humans. Remains of these plants in the cave deposits would likely be detected by both pollen and sedaDNA. Comparison of the pollen and sedaDNA records revealed six families and two genera in common. Of the two genera (Quercus and Artemisia), Quercus may be an unlikely pioneer plant of cave environments, but its presence cannot be excluded. Artemisia species are described as preferring no-shade to semi-shade conditions (Fern, 1997). With the cave opening towards the northeast, it enjoys only morning sun and therefore is unlikely to have sustained Artemisia growth.
We note a few instances of simultaneous detection of plant families by sedaDNA and pollen. Brassicaceae was recorded throughout AH VII and VI by sedaDNA, but occurred only in single pollen samples of AH VII, VI, and III. Both records indicate Amaranthaceae in AH VII, and VI, but only pollen of this family were identified in AH IV and III. Fagaceae was found throughout the pollen sequence, but only in two DNA samples. Plumbaginaceae was observed with sedaDNA throughout AH VI, but in only one pollen sample in AH VI. Lastly, Poaceae and Asteraceae were present in both pollen and sedaDNA in AH VII, VI, and V. However, Asteraceae was highly abundant in the pollen record in AH IV, while absent from the sedaDNA record of that layer.
Overall, we see a few layers where both records overlap. In contrast to the assumed higher potential of plants to grow at the cave entrance or inside the cave during periods of reduced occupation, we found the layers indicating overlap among proxies to be those associated with increased human occupation. For example, a lower abundance and diversity of plants was associated with the archaeologically sterile layers of AH V and IV (Figure 2, Figure 4). Previous palynological analysis, including investigation of nonpollen palynomorphs, also reported fewer other botanical remains in these sterile layers compared to the rich layers (Kandel et al., 2017). In sum, comparison of the sedaDNA and pollen proxies suggests the plant diversity we recovered at Aghitu-3 Cave was affected by animal or human occupation, while the effect of plants growing at the cave entrance or inside the cave on the recovered plant diversity was limited.
4.2. Reconstructed plant assemblages across archaeological horizons
Our analysis of pollen and plant sedaDNA in the sediment record of Aghitu-3 Cave revealed a diverse plant record with an abundance of Brassicaceae, Fabaceae, Amaranthaceae (supported by the sedaDNA), and Asteraceae (supported by the pollen data; SOM Figs. S1–S3). The diversity of plant taxa matched the total abundance patterns, with higher diversity in the lowest layers (AH VII and VI) and lower diversity in the almost sterile levels (AH V and IV; Fig. 2). For the pollen record, plant diversity also remained low in AH III, the level with the most archaeological finds, whereas for the sedaDNA record, plant diversity increased in AH III compared to AH V and IV (Fig. 2).
The reconstructed plant assemblages differed significantly among AHs, as supported by NMDS and perMANOVA (Fig. 3). The stratification in assemblages was stronger in the sedaDNA compared to the pollen, showing that the plant assemblages reconstructed using sedaDNA are more specific to the AHs than those identified using pollen. This may reflect an artifact of the nature of the sedaDNA data, with MOTUs providing a higher resolution than pollen types. Alternatively, this may indicate that the sedaDNA more closely reflects the delineation into archaeological horizons of human occupation alternating with sterile layers, where the presence of more or different plants resulted from increased human activity. The patterns in the reduced sedaDNA data set (species and genera only) support an association with human occupation in terms of diversity (Fig. 2A). The abundance and composition of the sedaDNA record match inferences from previous finds at Aghitu-3, with lithic densities indicating low settlement intensity in AH VII and VI, no significant occupation in AH V and IV, followed by high lithic density in AH III. This trend is mirrored by similar patterns in the presence of combustion features and evidence of animal exploitation (Kandel et al., 2017; Bertacchi et al., 2021).
The higher diversity in plant assemblages in AH VII, VI, and for sedaDNA also in AH III, matches changes in paleoclimate previously reconstructed from the deposition of the Aghitu-3 cave sediments based on micromammal and pollen data (Kandel et al., 2017). These proxies suggest that conditions were warm and humid during the deposition of AH VII and VI (∼39–32 kyr cal BP), while the climate was cooler and humid during AH V (∼32–30 kyr cal BP). Further cooling was seen during deposition of AH IV (∼30–29 kyr cal BP), with a cold and dry period in the lower part of AH III (∼29–26 kyr cal BP) followed by conditions that were not as cold during the upper part of AH III (∼26–24 kyr cal BP). These reconstructed environmental changes are in accordance with the global climatic trend for the period covered (Lisiecki and Raymo, 2005). The sedaDNA data also supports this with higher plant DNA diversity as well as higher concentrations of DNA from plant taxa that prefer moist soil and occur in temperate and warm regions (e.g., Atriplex sp., Brassica sp., Pisum sp.) in layers deposited during warmer periods (AHs VII and VI) compared to those deposited during colder periods (AHs V, IV, and AH III). During the upper part of AH III, conditions were not as cold as during the lower part of this layer and plant DNA diversity increased. Archaeological finds from this layer suggest high settlement intensity with high lithic density, the appearance of bone tools and shell beads, and frequent combustion features (Kandel et al., 2017). Despite intense site-use during AH III, plant DNA diversity did not return to the high levels found in the older human occupation layers (AHs VII and VI; Fig. 2A) and the plant assemblages are different (Fig. 3). This is likely due to the cold, dry conditions affecting the vegetation communities around the cave as well as the potential loss of sedimentary DNA by the increased fire activity in AH III, as heat degrades DNA.
Climatic conditions affect the distribution of plant and animal communities and can reasonably be assumed to influence the form and intensity of site occupation by humans (Avery, 1995; Armitage et al., 2011; Pinhasi et al., 2011; Scerri et al., 2014). Higher humidity especially facilitates the expansions of woody plants and a diversification of the landscape, as is the case today in more humid parts of the southern Caucasus (Gulisashvili et al., 1975). The warm and humid conditions during AHs VII and VI likely facilitated the establishment of a higher diversity of plants in the vicinity of the cave and may have attracted human occupation at Aghitu-3 Cave. Behavioral changes observed in AH III, illustrated by the frequent combustion features and the appearance of bone tools, including a needle and awl suggesting manufacturing of clothing (Kandel et al., 2017), presumably provided ways to cope with the harsher environmental conditions during this period.
4.3. Potential prehistoric human plant use at Aghitu-3
An assessment of prehistoric plant use is inherently based on our current relation with plants supplemented by traditional knowledge that has persisted. Of the 43 plant taxa that were recovered at Aghitu-3, all but five were found to be useful to humans (Table 1; Fig. 4). This high proportion of useful taxa suggests that the plant assemblage recovered from the most archaeologically rich horizons in Aghitu-3 cave are a result of human occupation. Owing to the limited recovery of plant taxa from the archaeologically sterile layers, and the detection of very few not useful plant taxa overall, it was not possible to test for a transition in the proportion of useful versus not useful plants between archaeologically rich and archaeologically sterile layers.
Plants for food and flavor Edible plants can be used as either bulk food or flavoring. We found the highest number of plant taxa that could be used as bulk food in AHs VII and VI, primarily from the vegetable families of Brassicaceae and Amaranthaceae (Table 1). The Brassicaceae family, also known as the mustard or cabbage family, includes many species that can be used as leafy vegetables, and many of our modern winter vegetables. Brassica oleracea is known as wild cabbage and its modern cultivars include several common foods such as broccoli, kale, Brussels sprouts, and cauliflower. The Amaranth family (Amaranthaceae) was represented by saltbush (Atriplex sp.) and goosefoot (Chenopodium sp.), and species of both genera can be eaten like spinach. Other detected bulk foods in AH VII and VI included maple (Acer sp.), French sorrel (R. scutatus), plume thistles (Cirsium sp.), and Pisum sp. (recently renamed to Lathyrus; Govaerts et al., 2021). One pollen sample in AH VI (at −4.26 m) with high plant richness further included B. vulgaris-type pollen, likely from common beet, as well as pollen from reed grasses (Phragmites sp.), Typha sp. and A. calamus. In Armenia, only one species of Phragmites (P. australis) is an accepted taxon in the Plants of the World Online database and all of its parts are edible; the stems have a high sugar content, and the roots are rich in starch. Typha has many common names including bulrush, reed, and cattail, and the underground storage organs are especially rich in starch as is also the case for A. calamus. Evidence of preserved starch grains on grinding stones indicates underground storage organs of Typha were eaten and processed for flour in Europe and the Russian plain up to 30,000 years ago (Revedin et al., 2010).
Like bulk food, more flavoring plant taxa were found in human occupation layers AH VII and VI compared to the other archaeological horizons. This is especially the case for the sedaDNA dataset (Fig. 4). All 11 flavoring plants detected through pollen were recorded in one sample (depth = −4.26 m), with several taxa limited to this sample, such as A. calamus, Knautia arvensis type, and Thalictrum sp. In contrast, pollen from pine trees (Pinus sp.) and oak (Quercus frainetto type) were observed throughout the stratigraphy (although pollen were generally detected in very low numbers), while pine DNA remained absent from the sedaDNA record and oak DNA was only documented in AHs VII and VI (SOM Fig. S2). Acorn pieces or meal can be used as a thickener in stews or for making bread. Roasted acorns have been used for making acorn coffee, but oak also has many other potential uses, including medicinal, repellent, ink, basketry, gum, as well as building material and fuel (Fern, 1997). Other plant taxa that were identified as flavoring include Artemisia and Astragalus, both large and diverse plant genera encompassing many species that can also be used for medicinal purposes.
Medicinal plants and diseases Most of the recovered plants were found to be of potential medicinal use (Fig. 4). Artemisia sp. is the most abundant plant in the pollen record with a high concentration in one sample in AH III and further detection in AHs VII, VI, and V. Its DNA was recorded in three samples in AHs VII and VI. Artemisia constitutes a diverse plant genus, of which 24 different species occur in Armenia (Table 1). Many of these have reported medicinal properties and are used against diseases such as malaria, inflammation, hepatitis, and for treating infections (Abad et al., 2012; Nigam et al., 2019). Artemisia annua has antimalarial properties, whereas Artemisia absinthium has been reported to have many different properties, including anti-inflammatory, antioxidant, antiseptic, and antimicrobial. Both of these species have been suggested to have been used as medicine by Upper Paleolithic humans in the Caucasus (Martkoplishvili and Kvavadze, 2015). However, as shown in Table 1, Artemisia species can also be used as flavoring, and as cave floor strewing for its fragrant smell and insect repellent properties. Similarly, the genus Astragalus—detected in most samples of AHs VI and III using sedaDNA—could have been used as flavoring, but is also known for its medicinal properties (Rios and Waterman, 1997; Auyeung et al., 2016).
A few detected taxa were limited to the category of medicinal plants and unlikely to be used for food or other purposes: Polemonium caeruleum, Buxus sempervirens, and Achillea pseudoaleppica. Both P. caeruleum (also known as Greek valerian and Jacob's ladder) and B. sempervirens were detected from pollen but not from sedaDNA, in AH IV and AHs IV and VI, respectively. Owing to the low concentrations of both plants at 1.5 up to 7.5 grains per gram of sediment and constituting less than 5% of the total pollen counts per sample, wind dispersal or other means of transfer onto the cave floor cannot be excluded. Polemonium caeruleum is a forb that prefers moist well-drained soil and has been used in the treatment of many conditions ranging from toothaches and headaches to fevers and epilepsy (Fern, 1997; Łaska et al., 2019). Buxus sempervirens, known as boxwood or common box, is an evergreen shrub and its leaves and bark have been used to treat various diseases. Tea made from the leaves can be used to reduce fever and has been reported to be drunk every day in Anatolia for its antiparasitic as well as its stimulating effects on bile and the liver (Baytop et al., 1999, cited by Orhan et al., 2012). Other traditional uses of this plant include as a laxative and purgative, a sleeping aid, sedative, diuretic, and for the treatment of malaria (Fern, 1997; Neves et al., 2009). DNA from Achillea pseudoaleppica was recorded in AHs VII and VI. This species occurs especially in steppic habitats on slopes and rocky surfaces. It is not edible, but Ac. pseudoaleppica and other members of the Achillea genus, commonly known as yarrow, are widely used in folk medicine, as the entire plant can be used for treating a variety of ailments. It is particularly valuable for treating wounds, colds, fevers, kidney diseases, stomach aches, as pain relief, and to reduce inflammation (Fern, 1997; Si et al., 2006; Nemeth and Bernath, 2008). Chemical analysis of essential oils of Ac. pseudoaleppica and other species of yarrow indicates high concentrations of anti-inflammatory and antimicrobial components (Özen et al., 2003; Si et al., 2006; Motavalizadehkakhky et al., 2013). Analyses at cave sites in Georgia have previously recovered pollen from common yarrow (Achillea millefolium) in Upper Paleolithic layers of Dzudzuana, Satsurblia, Kotias Klde, and Bondi caves (Martkoplishvili and Kvavadze, 2015). Pollen counts were high in the Upper Paleolithic layers compared to modern pollen spectra inside and outside several karst caves, suggesting that humans brought these plants into the caves (Martkoplishvili and Kvavadze, 2015). Combined with our findings this indicates that the use of yarrow for the treatment of ailments could have been a common practice across the region during the Upper Paleolithic.
Plants for various other uses Several recovered plants at Aghitu-3 Cave are known repellents and other potential uses included strewing (i.e., the scattering of fragrant or insect repellent plants), basketry, tinder, dyeing, and string (Table 1). Four of the 43 plant taxa recovered at Aghitu-3 can be used for making string, and archaeological finds at Aghitu-3 Cave include an eyed needle made of bone recovered from AH III (Kandel et al., 2017), suggesting the manufacturing of clothing. The oldest reported dyed textile fibers to date were found in Dzudzuana Cave in Georgia, made from wild flax and dated to approximately 30,000 years ago (Kvavadze et al., 2009). At Aghitu-3, we found that 11 of the 43 plant taxa recovered can be used as dye, for example, to color textiles. For instance, Isatis tinctoria is also known as dyer's woad for the indigo dye that can be produced from the leaves, and P. australis (common reed) can be used to make a light green dye from the inflorescence, while the sturdiness of the stems and leaves makes it a good material for constructions, fences, thatching, mats, nets, and rope (Fern, 1997). The detection of several plants that can be used to produce and dye textiles, together with the finding of an eyed needle at Aghitu-3 Cave and the previously reported dyed flax from the same region suggests that manufacturing and dyeing of textiles may have been common practice. Altogether, the recovery of a wide range of plant taxa that can be used for food, flavoring, medicine, and/or technical purposes, demonstrates the potential of the area surrounding Aghitu-3 to support Upper Paleolithic humans.
5. Conclusions
Survival of identifiable plant remains is rare at Paleolithic sites. Our current knowledge about human plant-use during this period is defined and limited by the degradation and recovery of plant remains and the availability of methods and technologies for plant detection and identification. Caves can provide unique environments in which the full effect of the elements on accumulated organic materials may be mitigated. Using evidence from sedaDNA and pollen, we were able to reconstruct the plant assemblages of the 15,000 yearlong sedimentary record of Aghitu-3 Cave.
The stratification of plant abundance and diversity differed among the previously described archaeological horizons. A lower abundance and diversity of plants was associated with the archaeologically sterile layers (AH V and IV). Especially sedaDNA reflected periods of human occupation with higher diversity detected in horizons with increased human activity (AH VII, VI, and III). The combination of low pollen concentrations with high sedaDNA abundance suggests animals or humans may have brought plant remains into the cave during AH VII and VI. This hypothesis is strengthened as most of the recovered plants are useful for food, flavoring, medicine, and/or other technical purposes. Moreover, we identified several specific taxa that support previous findings about human plant use (e.g., medicine, dye) in the region during the Upper Paleolithic. This study represents the first application of plant sedaDNA analysis on cave sediments for the investigation of potential plant use by prehistoric humans.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The excavation and research programs at Aghitu were funded by The Gfoeller Renaissance Foundation (USA), the Armenian National Academy of Sciences, and the Heidelberg Academy of Sciences and Humanities. Further financial support was provided by the Faculty of Mathematical and Natural Sciences, University of Oslo. We thank the Norwegian Sequencing Centre for their advice and DNA sequencing. DNA analyses were performed on the Saga computer cluster, owned by the University of Oslo and Uninett/Sigma2, and operated by the Department of Research Computing at USIT, University of Oslo.
Author contributions
DownloadDownload all supplementary files included with this article
HJB, SB, AWK, and ATMS, conceptualization; SB, AWK, AAB, and ATMS, design; BG and AWK, excavation; AWK and ATMS, sampling; ATMS, sedaDNA and bioinformatic analysis; AAB, pollen analysis; AAB, RWB, and AKB, validation of sedaDNA identifications; AAB, plant use assessment; and ATMS, statistical analysis and writing original draft. All co-authors commented on the manuscript.
Supplementary Online Material
The following are the Supplementary online material to this article:PDF DownloadDownload : Download Acrobat PDF file (630KB)
Multimedia component 1.
Interactive Case InsightsDownload : Download spreadsheet (210KB)
Multimedia component 2.
Recommended articles
References
- Abad et al., 2012
- M.J. Abad, L.M. Bedoya, L. Apaza, P. Bermejo
The Artemisia L. genus: A review of bioactive essential oils
Molecules, 17 (2012), pp. 2542-2566
- Albert et al., 2000
- R.M. Albert, S. Weiner, O. Bar-Yosef, L. Meignen
Phytoliths in the Middle Palaeolithic deposits of Kebara Cave, Mt Carmel, Israel: Study of the plant materials used for fuel and other purposes
J. Archaeol. Sci., 27 (2000), pp. 931-947
- Allué et al., 2012
- E. Allué, D. Cabanes, A. Solé, R. Sala
Hearth functioning and forest resource exploitation based on the archeobotanical assemblage from Level J
E.C. Roura (Ed.), High Resolution Archaeology and Neanderthal Behavior, Springer, Dordrecht (2012), pp. 373-385
- Alsos et al., 2018
- I.G. Alsos, Y. Lammers, N.G. Yoccoz, T. Jørgensen, P. Sjögren, L. Gielly, M.E. Edwards
Plant DNA metabarcoding of lake sediments: How does it represent the contemporary vegetation
PLoS One, 13 (2018), Article e0195403
- Altolaguirre et al., 2021
- Y. Altolaguirre, M. Schulz, L. Gibert, A.A. Bruch
Mapping Early Pleistocene environments and the availability of plant food as a potential driver of early Homo presence in the Guadix-Baza Basin (Spain)
J. Hum. Evol., 155 (2021), Article 102986
- Armitage et al., 2011
- S.J. Armitage, S.A. Jasim, A.E. Marks, A.G. Parker, V.I. Usik, H.P. Uerpmann
The southern route “out of Africa”: Evidence for an early expansion of modern humans into Arabia
Science, 331 (6016) (2011), pp. 453-456
- Auyeung et al., 2016
- K.K. Auyeung, Q.-B. Han, J.K. Ko
Astragalus membranaceus: A review of its protection against inflammation and gastrointestinal cancers
Am. J. Chin. Med., 44 (2016), pp. 1-22
- Avery, 1995
- D.M. Avery
Physical environment and site choice in South Africa
J. Archaeol. Sci., 22 (1995), pp. 343-353
- Bertacchi et al., 2021
- A. Bertacchi, B. Gasparyan, B. Gruwier, F. Rivals, A.W. Kandel
Upper Paleolithic animal exploitation in the Armenian Highlands: The zooarchaeology of Aghitu-3 Cave
Quat. Int., 587–588 (2021), pp. 400-414
- Birks and Bjune, 2010
- H.H. Birks, A.E. Bjune
Can we detect a west Norwegian tree line from modern samples of plant remains and pollen? Results from the DOORMAT project
Veg. Hist. Archaeobot., 19 (2010), pp. 325-340
- Bland and Altman, 1995
- J.M. Bland, D.G. Altman
Multiple significance tests: The Bonferroni method
BMJ, 310 (1995), p. 170
- Bohn et al., 2004
- U. Bohn, G. Gollub, C. Hettwer, Z. Neuhäuslová, T. Raus, H. Schlüter, H. Weber
Map of the Natural Vegetation of Europe
German Federal Agency for Nature Conservation, Bonn (2004)
- Boyer et al., 2016
- F. Boyer, C. Mercier, A. Bonin, Y. Le Bras, P. Taberlet, E. Coissac
Obitools: A unix-inspired software package for DNA metabarcoding
Mol. Ecol. Res., 16 (2016), pp. 176-182
- Bruch and Gabrielyan, 2002
- A.A. Bruch, I.G. Gabrielyan
Quantitative data of the Neogene climatic development in Armenia and Nakhichevan
Acta Univ. Carol. Geol., 46 (2002), pp. 41-48
- Clarke, 1993
- K.R. Clarke
Non-parametric multivariate analyses of changes in community structure
Austral Ecol., 18 (1993), pp. 117-143
- Clarke et al., 2020
- C.L. Clarke, I.G. Alsos, M.E. Edwards, A. Paus, L. Gielly, H. Haflidason, J. Mangerud, C. Regnéll, P.D.M. Hughes, J.I. Svendsen, A.E. Bjune
A 24,000-year ancient DNA and pollen record from the Polar Urals reveals temporal dynamics of arctic and boreal plant communities
Quat. Sci. Rev., 247 (2020), Article 106564
- Egeland et al., 2010
- C.P. Egeland, C.M. Nicholson, B. Gasparian
Using GIS and ecological variables to identify high potential areas for paleoanthropological survey: An example from Northern Armenia
J. Ecol. Anthropol., 14 (2010), pp. 89-98
- Ellis et al., 2008
- J.R. Ellis, K.E. Bentley, D.E. McCauley
Detection of rare paternal chloroplast inheritance in controlled crosses of the endangered sunflower Helianthus verticillatus
Heredity, 100 (2008), pp. 574-580
- Epp et al., 2019
- L.S. Epp, H.H. Zimmermann, K.R. Stoof-Leichsenring
Sampling and extraction of ancient DNA from sediments
B. Shapiro, A. Barlow, P.D. Heintzman, M. Hofreiter, J.L.A. Paijmans, A.E.R. Soares (Eds.), Ancient DNA, Humana Press, New York (2019), pp. 31-44
- Fayvush and Aleksanyan, 2016
- G.M. Fayvush, A.S. Aleksanyan
Habitats of Armenia
Botanical Institute of Armenian Academy of Sciences, Yerevan (2016)
- Fayvush et al., 2017
- G. Fayvush, A. Aleksanyan, R.W. Bussmann
Ethnobotany of the Caucasus – Armenia
R.W. Bussmann (Ed.), Ethnobotany of the Caucasus, Springer International Publishing, Cham (2017)
- Fayvush et al., 2013
- G. Fayvush, K. Tamanyan, M. Kalashyan, E. Vitek
“Biodiversity Hotspots” in Armenia
Annal. Naturhist. Mus. Wien, B Bot. Zool. (2013), pp. 11-20
- Fern, 1997
- K. Fern
Plants for a Future: Edible and Useful Plants for a Healthier World
Permanent Publications (1997)
- Fern, 2019
- K. Fern
Useful temperate plants
(2019)
- Ferring et al., 2011
- R. Ferring, O. Oms, J. Agustí, F. Berna, M. Nioradze, T. Shelia, M. Tappen, A. Vekua, D. Zhvania, D. Lordkipanidze
Earliest human occupations at Dmanisi (Georgian Caucasus) dated to 1.85–1.78 Ma
Proc. Natl. Acad. Sci. USA, 108 (2011), pp. 10432-10436
- Ficetola et al., 2010
- G. Ficetola, E. Coissac, S. Zundel, T. Riaz, W. Shehzad, J. Bessière, P. Taberlet, F. Pompanon
An In silico approach for the evaluation of DNA barcodes
BMC Genom., 11 (2010), p. 434
- Fleischhauer et al., 2016
- S.G. Fleischhauer, J. Guthmann, R. Spiegelberger
Enzyklopädie Essbare Wildpflanzen
AT Verlag, Aarau (2016)
- Gabrielyan and Gasparyan, 2003
- I.G. Gabrielyan, B. Gasparyan
Usloviya prozhivaniya pervobitnogo cheloveka v kanyone reki Vorotan po paleontologicheskim dannim
A. Kalantaryan (Ed.), Arxeologiya, Etnologiya i Folkloristika Kavkaza, Materiali Mezhdunarodnoy Konferentsii (2003), pp. 23-29
- Gabunia and Vekua, 1995
- L. Gabunia, A. Vekua
A Plio-Pleistocene hominid from Dmanisi, East Georgia, Caucasus
Nature, 373 (1995), pp. 509-512
- Gasparyan et al., 2014
- B. Gasparyan, A.W. Kandel, C. Montoya
Living the high life: The Upper Paleolithic Settlement of the Armenian Highlands
B. Gasparyan, M. Arimura (Eds.), Stone Age of Armenia, A Guide-book to the Stone Age Archaeology in the Republic of Armenia, Kanazawa University Press, Japan (2014), pp. 107-131
- Gasparyan and Glauberman, 2022
- B. Gasparyan, P. Glauberman
Beyond European boundaries: Neanderthals in the Armenian Highlands and the Caucasus
F. Romagnoli, F. Rivals, S. Benazzi (Eds.), Updating Neanderthals: Understanding Behavioral Complexity in the Late Middle Paleolithic, Academic Press, Elsevier (2022), pp. 275-301
- Giguet-Covex et al., 2019
- C. Giguet-Covex, G.F. Ficetola, K. Walsh, J. Poulenard, M. Bajard, L. Fouinat, P. Sabatier, L. Gielly, E. Messager, A.L. Develle, F. David, P. Taberlet, E. Brisset, F. Guiter, R. Sinet, F. Arnaud
New insights on lake sediment DNA from the catchment: Importance of taphonomic and analytical issues on the record quality
Sci. Rep., 9 (2019), Article 14676
- Goldberg et al., 2012
- P. Goldberg, H. Dibble, F. Berna, D. Sandgathe, S.J. McPherron, A. Turq
New evidence on Neandertal use of fire: Examples from Roc de Marsal and Pech de l'Azé IV
Quat. Int., 247 (2012), pp. 325-340
- Govaerts et al., 2021
- R. Govaerts, E. Nic Lughadha, N. Black, R. Turner, A. Paton
The World Checklist of Vascular Plants, a continuously updated resource for exploring global plant diversity
Sci. Data, 8 (2021), p. 215
- Gulisashvili et al., 1975
- V. Gulisashvili, L. Makhatadze, M. Prilipko
Rastitelnost Kavkaza (Vegetation of the Caucasus)
Nauka, Moscow (1975)
- Haile et al., 2007
- J. Haile, R. Holdaway, K. Oliver, M. Bunce, M.T.P. Gilbert, R. Nielsen, K. Munch, S.Y.W. Ho, B. Shapiro, E. Willerslev
Ancient DNA chronology within sediment deposits: Are paleobiological reconstructions possible and is DNA leaching a factor?
Mol. Ecol. Evol., 24 (2007), pp. 982-989
- Haouchar et al., 2014
- D. Haouchar, J. Haile, M.C. McDowell, D.C. Murray, N.E. White, R.J.N. Allcock, M.J. Phillips, G.J. Prideaux, M. Bunce
Thorough assessment of DNA preservation from fossil bone and sediments excavated from a late Pleistocene–Holocene cave deposit on Kangaroo Island, South Australia
Quat. Sci. Rev., 84 (2014), pp. 56-64
- Hardy, 2018
- K. Hardy
Plant use in the Lower and Middle Palaeolithic: Food, medicine and raw materials
Quat. Sci. Rev., 191 (2018), pp. 393-405
- Hardy, 2019
- K. Hardy
Paleomedicine and the use of plant secondary compounds in the Paleolithic and Early Neolithic
Evol. Anthropol. Issues News Rev., 28 (2019), pp. 60-71
- Hardy et al., 2012
- K. Hardy, S. Buckley, M.J. Collins, A. Estalrrich, D. Brothwell, L. Copeland, A. García-Tabernero, S. García-Vargas, M. De La Rasilla, C. Lalueza-Fox, R. Huguet, M. Bastir, D. Santamaría, M. Madella, J. Wilson, Á. Fernández Cortés, A. Rosas
Neanderthal medics? Evidence for food, cooking, and medicinal plants entrapped in dental calculus
Naturwissenschaften, 99 (2012), pp. 617-626
- Hofreiter et al., 2003
- M. Hofreiter, J.I. Mead, P. Martin, H.N. Poinar
Molecular caving
Curr. Biol., 13 (2003), pp. R693-R695
- Holm, 1979
- S. Holm
A simple sequentially rejective multiple test procedure
Scand. J. Stat., 6 (1979), pp. 65-70
- Hunt and Fiacconi, 2018
- C.O. Hunt, M. Fiacconi
Pollen taphonomy of cave sediments: What does the pollen record in caves tell us about external environments and how do we assess its reliability?
Quat. Int., 485 (2018), pp. 68-75
- Joannin et al., 2010
- S. Joannin, J.-J. Cornée, P. Münch, M. Fornari, I. Vasiliev, W. Krijgsman, S. Nahapetyan, I. Gabrielyan, V. Ollivier, P. Roiron, C. Chataigner
Early Pleistocene climate cycles in continental deposits of the Lesser Caucasus of Armenia inferred from palynology, magnetostratigraphy, and 40Ar/39Ar dating
Earth Planet Sci. Lett., 291 (2010), pp. 149-158
- Jørgensen et al., 2012
- T. Jørgensen, J. Haile, P. Möller, A. Andreev, S. Boessenkool, M. Rasmussen, F. Kienast, E. Coissac, P. Taberlet, C. Brochmann, N.H. Bigelow, K. Andersen, L. Orlando, M.T.P. Gilbert, E. Willerslev
A comparative study of ancient sedimentary DNA, pollen and macrofossils from permafrost sediments of northern Siberia reveals long-term vegetational stability
Mol. Ecol., 21 (2012), pp. 1989-2003
- Juggins and Juggins, 2020
- S. Juggins, M.S. Juggins
Package ‘rioja’
https://cran.r-project.org/web/packages/rioja/index.html (2020)
- Kandel et al., 2017
- A.W. Kandel, B. Gasparyan, E. Allué, G. Bigga, A.A. Bruch, V.L. Cullen, E. Frahm, R. Ghukasyan, B. Gruwier, F. Jabbour, C.E. Miller, A. Taller, V. Vardazaryan, D. Vasilyan, L. Weissbrod
The earliest evidence for Upper Paleolithic occupation in the Armenian Highlands at Aghitu-3 Cave
J. Hum. Evol., 110 (2017), pp. 37-68
- Kandel et al., 2014
- A.W. Kandel, B. Gasparyan, S. Nahapetyan, A. Taller, L. Weissbrod
The Upper Paleolithic settlement of the Armenian Highlands. Modes de Contacts et de Déplacements Au Paléolithique Eurasiatique—Modes of Contact and Displacements during the Eurasian Palaeolithic
Actes Du Colloque International de La Commission, 8 (2014), pp. 39-62
- Kassambara, 2021
-
A. Kassambara
rstatix: Pipe-friendly framework for basic statistical tests [Manual]
- Kvavadze et al., 2009
- E. Kvavadze, O. Bar-Yosef, A. Belfer-Cohen, E. Boaretto, N. Jakeli, Z. Matskevich, T. Meshveliani
30,000-year-old wild flax fibers
Science, 325 (5946) (2009), p. 1359
- Łaska et al., 2019
- G. Łaska, E. Sieniawska, Ł. Świątek, J. Zjawiony, S. Khan, A. Boguszewska, M. Stocki, M. Angielczyk, M. Polz-Dacewicz
Phytochemistry and biological activities of Polemonium caeruleum L
Phytochem. Lett., 30 (2019), pp. 314-323
- Li, 2018
- D. Li
HillR: Taxonomic, functional, and phylogenetic diversity and similarity through Hill Numbers
J. Open Source Softw., 3 (31) (2018), p. 1041
- Lisiecki and Raymo, 2005
- L.E. Lisiecki, M.E. Raymo
A Pliocene-Pleistocene stack of 57 globally distributed benthic δ18O records
Paleoceanography, 20 (2005), p. PA1003
- Lyubin and Belyaeva, 2013
- V.P. Lyubin, E.V. Belyaeva
Paleolithic Armenia
H.Y. Simonyan (Ed.), Archaeological Heritage of Armenia, Hushardzan Publishers, Yerevan (2013), pp. 14-17
- Martkoplishvili and Kvavadze, 2015
- I. Martkoplishvili, E. Kvavadze
Some popular medicinal plants and diseases of the Upper Palaeolithic in Western Georgia
J. Ethnopharmacol., 166 (2015), pp. 42-52
- Massilani et al., 2022
- D. Massilani, M.W. Morley, S.M. Mentzer, V. Aldeias, B. Vernot, C. Miller, M. Stahlschmidt, M.B. Kozlikin, M.V. Shunkov, N.J.C. Derevianko, N.J. Conard, S. Wurz, C.S. Henshilwood, J. Vasquez, E. Essel, S. Nagel, J. Richter, B. Nickel, R.G. Roberts, S. Pääbo, V. Slon, P. Goldberg, M. Meyer
Microstratigraphic preservation of ancient faunal and hominin DNA in Pleistocene cave sediments
Proc. Natl. Acad. Sci. USA, 119 (2022), Article e2113666118
- Moreno-Sanchez and Sayadyan, 2005
- R. Moreno-Sanchez, H.Y. Sayadyan
Evaluation of the forest cover in Armenia
Int. For. Rev., 7 (2005), pp. 113-127
- Motavalizadehkakhky et al., 2013
- A. Motavalizadehkakhky, A. Shafaghat, H.A. Zamani, H. Akhlaghi, M. Mohammadhosseini, J. Mehrzad, Z. Ebrahimi
Compositions and the in vitro antimicrobial activities of the essential oils and extracts of two Achillea species from Iran
J. Med. Plant Res., 7 (2013), pp. 1280-1292
- Nakhutsrishvili, 2012
- G. Nakhutsrishvili
The vegetation of Georgia (South Caucasus)
Springer-Verlag Berlin Heidelberg (2012)
- Nemeth and Bernath, 2008
- E. Nemeth, J. Bernath
Biological activities of yarrow species (Achillea spp.)
Curr. Pharm. Des., 14 (2008), pp. 3151-3167
- Neves et al., 2009
- J.M. Neves, C. Matos, C. Moutinho, G. Queiroz, L.R. Gomes
Ethnopharmacological notes about ancient uses of medicinal plants in Trás-os-Montes (northern of Portugal)
J. Ethnopharmacol., 124 (2009), pp. 270-283
- Niemeyer et al., 2017
- B. Niemeyer, L.S. Epp, K.R. Stoof-Leichsenring, L.A. Pestryakova, U. Herzschuh
A comparison of sedimentary DNA and pollen from lake sediments in recording vegetation composition at the Siberian treeline
Mol. Ecol. Res., 17 (2017), pp. e46-e62
- Nigam et al., 2019
- M. Nigam, M. Atanassova, A.P. Mishra, R. Pezzani, H.P. Devkota, S. Plygun, B. Salehi, W.N. Setzer, J. Sharifi-Rad
Bioactive compounds and health benefits of Artemisia species
Nat. Prod. Commun., 14 (2019), p. 7
- Oksanen et al., 2020
-
J. Oksanen, F.G. Blanchet, M. Friendly, R. Kindt, P. Legendre, D. McGlinn, P.R. Minchin, R.B. O'Hara, G.L. Simpson, P. Solymos, M.H.H. Stevens, E. Szoecs, H. Wagner
vegan: Community ecology package [Manual]
- Ollivier et al., 2010
- V. Ollivier, S. Nahapetyan, P. Roiron, I. Gabrielyan, B. Gasparyan, C. Chataigner, S. Joannin, J.-J. Cornée, H. Guillou, S. Scaillet, P. Munch, W. Krijgsman
Quaternary volcano-lacustrine patterns and palaeobotanical data in southern Armenia
Quat. Int., 223 (2010), pp. 312-326
- Orhan et al., 2012
- I.E. Orhan, S.A. Erdem, F.S. Senol, M. Kartal, B. Sener
Exploration of cholinesterase and tyrosinase inhibitory, antiprotozoal and antioxidant effects of Buxus sempervirens L. (boxwood)
Ind. Crops Prod., 40 (2012), pp. 116-121
- Özen et al., 2003
- H.Ç. Özen, Z. Toker, R.A. Clery, N.E. Owen
Composition of the essential oil of Achillea pseudoaleppica Hub.-Mor
J. Essent. Oil Res., 15 (2003), pp. 96-97
- Parducci et al., 2017
- L. Parducci, K.D. Bennett, G.F. Ficetola, I.G. Alsos, Y. Suyama, J.R. Wood, M.W. Pedersen
Ancient plant DNA in lake sediments
New Phytol., 214 (2017), pp. 924-942
- Parducci et al., 2018
- L. Parducci, K. Nota, J. Wood
Reconstructing past vegetation communities using ancient DNA from lake sediments
C. Lindqvist, O.P. Rajora (Eds.), Paleogenomics, Springer, Cham (2018), pp. 163-187
- Pedersen et al., 2015
- M.W. Pedersen, S. Overballe-Petersen, L. Ermini, C.D. Sarkissian, J. Haile, M. Hellstrom, J. Spens, P.F. Thomsen, K. Bohmann, E. Cappellini, I. Bærholm Schnell, N.A. Wales, C. Carøe, P.F. Campos, A.M.Z. Schmidt, M.T.P. Gilbert, A.J. Hansen, L. Orlando, E. Willerslev
Ancient and modern environmental DNA
Philos. Trans. R. Soc. Lond. B Biol. Sci., 370 (2015), Article 20130383
- Pinhasi et al., 2011
- R. Pinhasi, B. Gasparian, S. Nahapetyan, G. Bar-Oz, L. Weissbrod, A.A. Bruch, R. Hovsepyan, K. Wilkinson
Middle Palaeolithic human occupation of the high altitude region of Hovk-1, Armenia
Quat. Sci. Rev., 30 (2011), pp. 3846-3857
- Power et al., 2018
- R.C. Power, D.C. Salazar-García, M. Rubini, A. Darlas, K. Harvati, M. Walker, J.-J. Hublin, A.G. Henry
Dental calculus indicates widespread plant use within the stable Neanderthal dietary niche
J. Hum. Evol., 119 (2018), pp. 27-41
- R Core Team, 2021
- R Core Team
R: A language and environment for statistical computing [Manual]
(2021)
- Revedin et al., 2010
- A. Revedin, B. Aranguren, R. Becattini, L. Longo, E. Marconi, M.M. Lippi, N. Skakun, A. Sinitsyn, E. Spiridonova, J. Svoboda
Thirty thousand-year-old evidence of plant food processing
Proc. Natl. Acad. Sci. USA, 107 (2010), pp. 18815-18819
- Rios and Waterman, 1997
- J.L. Rios, P.G. Waterman
A review of the pharmacology and toxicology of Astragalus
Phytother. Res., 11 (1997), pp. 411-418
- Rivera et al., 2012
-
D. Rivera, G. Matilla, C. Obón, F. Alcaraz
Plants and Humans in the Near East and the Caucasus: Ancient and Traditional Uses of Plants as Food and Medicine, a Diachronic Ethnobotanical Review, s. vols. 1–2, Ediciones de la Universidad de Murcia (2012)
- Rohland et al., 2018
- N. Rohland, I. Glocke, A. Aximu-Petri, M. Meyer
Extraction of highly degraded DNA from ancient bones, teeth and sediments for high-throughput sequencing
Nat. Protoc., 13 (2018), pp. 2447-2461
- Salazar-García et al., 2013
- D.C. Salazar-García, R.C. Power, A.S. Serra, V. Villaverde, M.J. Walker, A.G. Henry
Neanderthal diets in central and southeastern Mediterranean Iberia
Quat. Int., 318 (2013), pp. 3-18
- Scerri et al., 2014
- E.M. Scerri, N.A. Drake, R. Jennings, H.S. Groucutt
Earliest evidence for the structure of Homo sapiens populations in Africa
Quat. Sci. Rev., 101 (2014), pp. 207-216
- Seersholm et al., 2020
- F.V. Seersholm, D.J. Werndly, A. Grealy, T. Johnson, E.M.K. Early, E.L. Lundelius, B. Winsborough, G.E. Farr, R. Toomey, A.J. Hansen, B. Shapiro, M.R. Waters, G. McDonald, A. Linderholm, T.W. Stafford Jr., M. Bunce
Rapid range shifts and megafaunal extinctions associated with late Pleistocene climate change
Nat. Commun., 11 (2020), p. 2770
- Shipley and Kindscher, 2016
- G.P. Shipley, K. Kindscher
Evidence for the paleoethnobotany of the Neanderthal: A review of the literature
Scientifica, 2016 (2016), Article 8927654
- Si et al., 2006
- X.-T. Si, M.-L. Zhang, Q.-W. Shi, H. Kiyota
Chemical constituents of the plants in the genus Achillea
Chem. Biodivers., 3 (2006), pp. 1163-1180
- Sjögren et al., 2017
- P. Sjögren, M.E. Edwards, L. Gielly, C.T. Langdon, I.W. Croudace, M.K.F. Merkel, T. Fonville, I.G. Alsos
Lake sedimentary DNA accurately records 20th Century introductions of exotic conifers in Scotland
New Phytol., 213 (2017), pp. 929-941
- Slon et al., 2017
- V. Slon, C. Hopfe, C.L. Weiß, F. Mafessoni, M. de la Rasilla, C. Lalueza-Fox, A. Rosas, M. Soressi, M.V. Knul, R. Miller, J.R. Stewart, A.P. Derevianko, Z. Jacobs, B. Li, R.G. Roberts, M.V. Shunkov, H. de Lumley, C. Perrenoud, I. Gušić, Ž. Kućan, P. Rudan, A. Aximu-Petri, E. Essel, S. Nagel, B. Nickel, A. Schmidt, K. Prüfer, J. Kelso, H.A. Burbano, S. Pääbo, M. Meyer
Neandertal and Denisovan DNA from Pleistocene sediments
Science, 356 (2017), pp. 605-608
- Stahlschmidt et al., 2019
- M.C. Stahlschmidt, T. Collin, D. Fernandes, G. Bar-Oz, A. Belfer-Cohen, Z. Gao, N. Jakeli, Z. Matskevich, T. Meshveliani, J. Pritchard, F. McDermott, R. Pinhasi
Ancient mammalian and plant DNA from late Quaternary stalagmite layers at Solkota Cave, Georgia
Sci. Rep., 9 (2019), p. 6628
- Stockmarr, 1971
- J. Stockmarr
Tablets with spores used in absolute pollen analysis
Pollen Spores, 13 (1971), pp. 615-621
- Taberlet et al., 2007
- P. Taberlet, E. Coissac, F. Pompanon, L. Gielly, C. Miquel, A. Valentini, T. Vermat, G. Corthier, C. Brochmann, E. Willerslev
Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding
Nucl. Acids Res., 35 (2007), p. e14
- Takhtajan, 1954
-
A.L. Takhtajan
Flora Armenii, s. vols. 1–11, Institute of Botany of the Armenian National Academy of Sciences, Yerevan (1954)
- Taller et al., 2018
- A. Taller, B. Gasparyan, A.W. Kandel
Living on the edge: The earliest modern human settlement of the Armenian Highlands in Aghitu-3 Cave
Y. Nishiaki, T. Akazawa (Eds.), The Middle and Upper Paleolithic Archeology of the Levant and Beyond, Springer, Singapore (2018), pp. 119-131
- Torti et al., 2015
- A. Torti, M.A. Lever, B.B. Jørgensen
Origin, dynamics, and implications of extracellular DNA pools in marine sediments
Mar. Genomics, 24 (2015), pp. 185-196
- Vernot et al., 2021
- B. Vernot, E.I. Zavala, A. Gómez-Olivencia, Z. Jacobs, V. Slon, F. Mafessoni, F. Romagné, A. Pearson, M. Petr, N. Sala, A. Pablos, A. Aranburu, J.M. Bermúdez de Castro, E. Carbonell, B. Li, M.T. Krajcarz, A.I. Krivoshapkin, K.A. Kolobova, M.B. Kozlikin, M.V. Shunkov, A.P. Derevianko, B. Viola, S. Grote, E. Essel, D.L. Herráez, S. Nagel, B. Nickel, J. Richter, A. Schmidt, B. Peter, J. Kelso, R.G. Roberts, J. Arsuaga, M. Meyer
Unearthing Neanderthal population history using nuclear and mitochondrial DNA from cave sediments
Science, 372 (2021), Article eabf1667
- Voldstad et al., 2020
- L.H. Voldstad, I.G. Alsos, W.R. Farnsworth, P.D. Heintzman, L. Håkansson, S.E. Kjellman, A. Rouillard, A. Schomacker, P.B. Eidesen
A complete Holocene lake sediment ancient DNA record reveals long-standing high Arctic plant diversity hotspot in northern Svalbard
Quat. Sci. Rev., 234 (2020), Article 106207
- Wadley et al., 2020
- L. Wadley, L. Backwell, F. d'Errico, C. Sievers
Cooked starchy rhizomes in Africa 170 thousand years ago
Science, 367 (2020), pp. 87-91
- Weyrich et al., 2017
- L.S. Weyrich, S. Duchene, J. Soubrier, L. Arriola, B. Llamas, J. Breen, A.G. Morris, K.W. Alt, D. Caramelli, V. Dresely, M. Farrell, A.G. Farrer, M. Francken, N. Gully, W. Haak, K. Hardy, K. Harvati, P. Held, E.C. Holmes, J. Kaidonis, C. Lalueza-Fox, M. de la Rasilla, A. Rosas, P. Semal, A. Soltysiak, G. Townsend, D. Usai, J. Wahl, D.H. Huson, K. Dobney, A. Cooper
Neanderthal behaviour, diet, and disease inferred from ancient DNA in dental calculus
Nature, 544 (2017), pp. 357-361
- Willerslev, 2003
- E. Willerslev
Diverse plant and animal genetic records from Holocene and Pleistocene sediments
Science, 300 (5620) (2003), pp. 791-795
- Willerslev and Cooper, 2005
- E. Willerslev, A. Cooper
Ancient DNA
Proc. R. Soc. B, 272 (2005), pp. 3-16
- Zavala et al., 2021
- E.I. Zavala, Z. Jacobs, B. Vernot, M.V. Shunkov, M.B. Kozlikin, A.P. Derevianko, E. Essel, C. de Fillipo, S. Nagel, J. Richter, F. Romagné, A. Schmidt, B. Li, K. O'Gorman, V. Slon, J. Kelso, S. Pääbo, R.G. Roberts, M. Meyer
Pleistocene sediment DNA reveals hominin and faunal turnovers at Denisova Cave
Nature, 565 (2021), pp. 399-403
- Zhang et al., 2020
- D. Zhang, H. Xia, F. Chen, B. Li, V. Slon, T. Cheng, R. Yang, Z. Jacobs, Q. Dai, D. Massilani, X. Shen, J. Wang, X. Feng, P. Cao, M.A. Yang, J. Yao, J. Yang, D.B. Madsen, Y. Han, W. Ping, F. Liu, C. Perreault, X. Chen, M. Meyer, J. Kelso, S. Pääbo, Q. Fu
Denisovan DNA in late Pleistocene sediments from Baishiya Karst Cave on the Tibetan Plateau
Science, 370 (2020), pp. 584-587
- Zhang et al., 2003
- Q. Zhang, Y. Liu, Sodmergen
Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cytoplasmic inheritance in 295 angiosperm species
Plant Cell Physiol., 44 (2003), pp. 941-951
Cited by (0)
© 2022 The Author(s). Published by Elsevier Ltd.